The rapidly advancing genome editing technologies have served as important tools for deciphering gene functions in complicated developmental processes. However, the application of these technologies in zebrafish still has some limitations. For example, the whole-embryo gene knockout with CRISPR/Cas9 technology often leads to early embryonic lethality, whereas the Cre/loxP conditional knockout system in zebrafish is restricted by the low knock-in germline transmission rate and knock-in site restrictions. These issues have partly hampered in-depth investigation of the spatiotemporal developmental regulation mechanisms in zebrafish.
On January 2, 2026, Anming Meng's lab from the School of Life Sciences at Tsinghua University published a study titled "Stage- and tissue-specific gene editing using 4-OHT-inducible Cas9 in whole organism" in the Journal of Cell Biology. This study developed a germline-specific inducible genome editing system in zebrafish and demonstrated that with 4-OHT treatment, the target genes in the zebrafish germline could be specifically knocked out at different developmental stages, achieving stage- and tissue-specific dual regulation. Furthermore, this inducible Cas9 protein could also achieve conditional gene knockout in early mouse embryos, presenting it as a potential tool for cross-species research.
The researchers constructed a 4-OHT-inducible Cas9 protein nCas9ERT2 based on 4-hydroxytamoxifen (4-OHT) and a mutated ligand-binding domain of estrogen receptor (ERT2), and validated that the nCas9ERT2 protein could be induced to translocate into nuclei and edit the target gene after 4-OHT treatment in the HEK293T cell line. Then, the researchers constructed a transgenic zebrafish line Tg(piwil1:nCas9ERT2-n3U) with germline-specific expression of nCas9ERT2 protein and a transgenic zebrafish line Tg(U6:gRNAs;cryaa:CFP) ubiquitously expressed gRNA of target genes. The experimental results showed that the inducible nCas9ERT2 protein could successfully respond to 4-OHT induction in both the primordial germ cells (PGCs) of zebrafish embryos and the oocytes of female zebrafish, achieving stage- and tissue-specific genome editing. Compared to traditional tissue-specific gene editing systems, this system effectively avoids off-target editing of early embryonic somatic cells by managing the timing of gene editing, highlighting its significant value for studying PGCs in early zebrafish embryos. Using this system, the researchers further explored the function of the tbx16 gene in PGC migration and found that the PGC-specific knockout of tbx16 would impair the PGC migration process, providing new experimental evidence for understanding the function of tbx16 in zebrafish embryonic development and expanding the knowledge of its regulatory role in the zebrafish PGC development process.
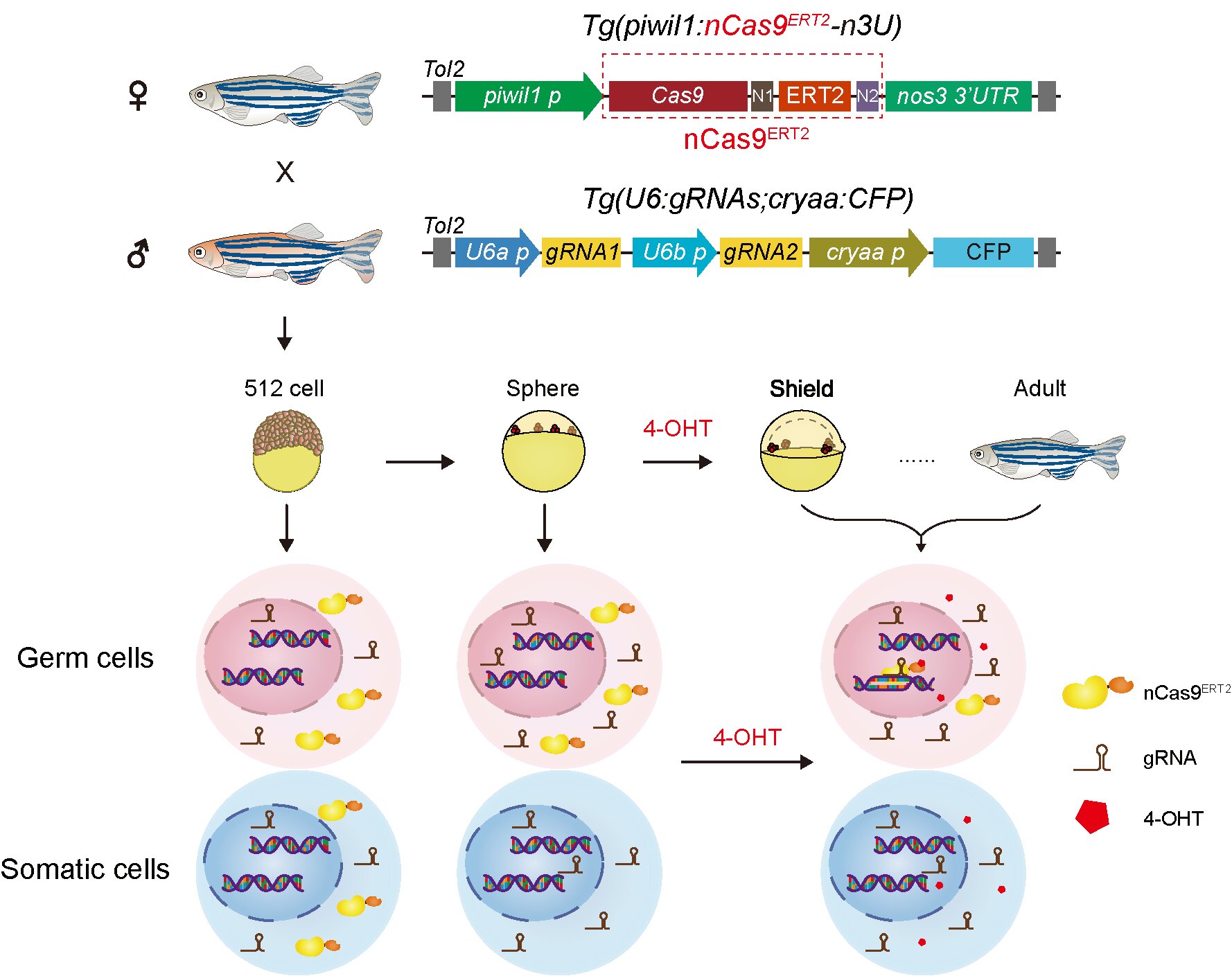
The schematic diagram of the zebrafish spatiotemporally inducible genome editing system and transgenic zebrafish lines.
Associate Researcher Xiaotong Wu and Academician Anming Meng from the School of Life Sciences, Tsinghua University, are the corresponding authors of this paper. Yaqi Li (Ph.D. graduate class of 2024), Weiying Zhang, and Zihang Wei (Ph.D. graduate class of 2025) are the co-first authors. Ph.D. candidates Han Li and Tursunjan Aziz, along with postdoctoral researchers Xin Liu and Tao Zheng, made significant contributions to this study. Associate Professor Cencan Xing from the University of Science and Technology Beijing participated in project guidance. This research was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China, the Tsinghua University-Peking University Joint Center for Life Sciences, and the State Key Laboratory of Membrane Biology.
Link: https://doi.org/10.1083/jcb.202412216
