In recent years, RNA has rapidly emerged as an important target in drug discovery. Accumulating evidence indicates that RNA is not merely a carrier of genetic information, but can also fold into complex and well-defined three-dimensional structures and engage in highly specific “lock-and-key” interactions with small molecules, thereby regulating critical biological processes. The successful approval of risdiplam, the first RNA-targeted small molecule drug for the treatment of spinal muscular atrophy, has further underscored the feasibility and therapeutic potential of this strategy. Nevertheless, traditional RNA-targetd drug discovery remains hampered by long development timelines, high costs, and low success rates, highlighting the urgent need for transformative approaches enabled by artificial intelligence and other emerging methodologies.
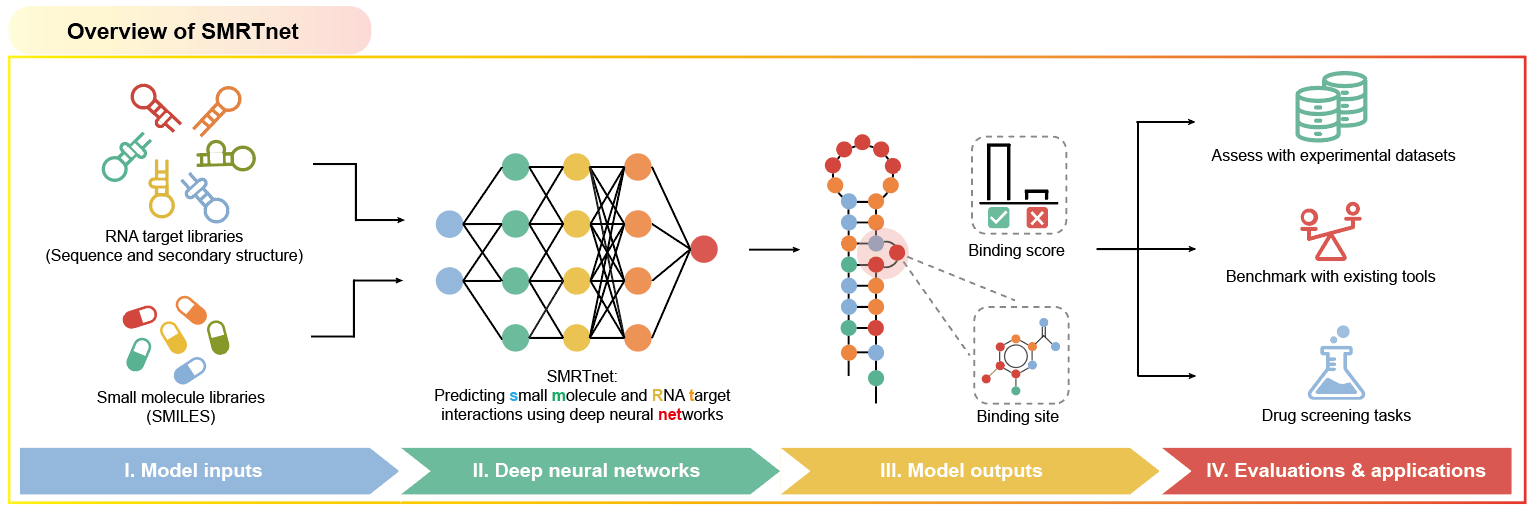
On January 2, 2026, a collaborative team led by Qiangfeng Cliff Zhang at Tsinghua University and Yangming Wang at Peking University reported a study entitled Predicting small molecule–RNA interactions without RNA tertiary structures in Nature Biotechnology. In this work, the authors developed an artificial intelligence (AI) method termed SMRTnet, which enables accurate prediction of small molecule–RNA interactions and their potential binding sites using only RNA secondary structure information. Through systematic computational analyses combined with experimental validation, the study comprehensively demonstrated the superior performance of SMRTnet in drug screening, providing a novel AI method for RNA therapeutics discovery.
The research team demonstrated that predictions generated by SMRTnet are highly consistent with known small molecule–RNA interactions, and that its overall predictive accuracy significantly outperforms existing state-of-the-art computational methods. Notably, SMRTnet also exhibits strong model interpretability, enabling accurate identification of potential small molecule binding sites on RNA. The authors further performed large-scale virtual screening followed by in vitro validation using a compound library comprising 7,350 natural products and metabolites across 10 disease-related RNA targets. Among the 190 candidate compounds prioritized by SMRTnet, the average experimental hit rate reached 21.1%, with hit rates for certain targets as high as 50%, far exceeding the ~0.1%–1% typically observed in conventional high-throughput screening. Collectively, these results demonstrate that SMRTnet can substantially reduce experimental costs and effectively accelerate early-stage drug screening and discovery.
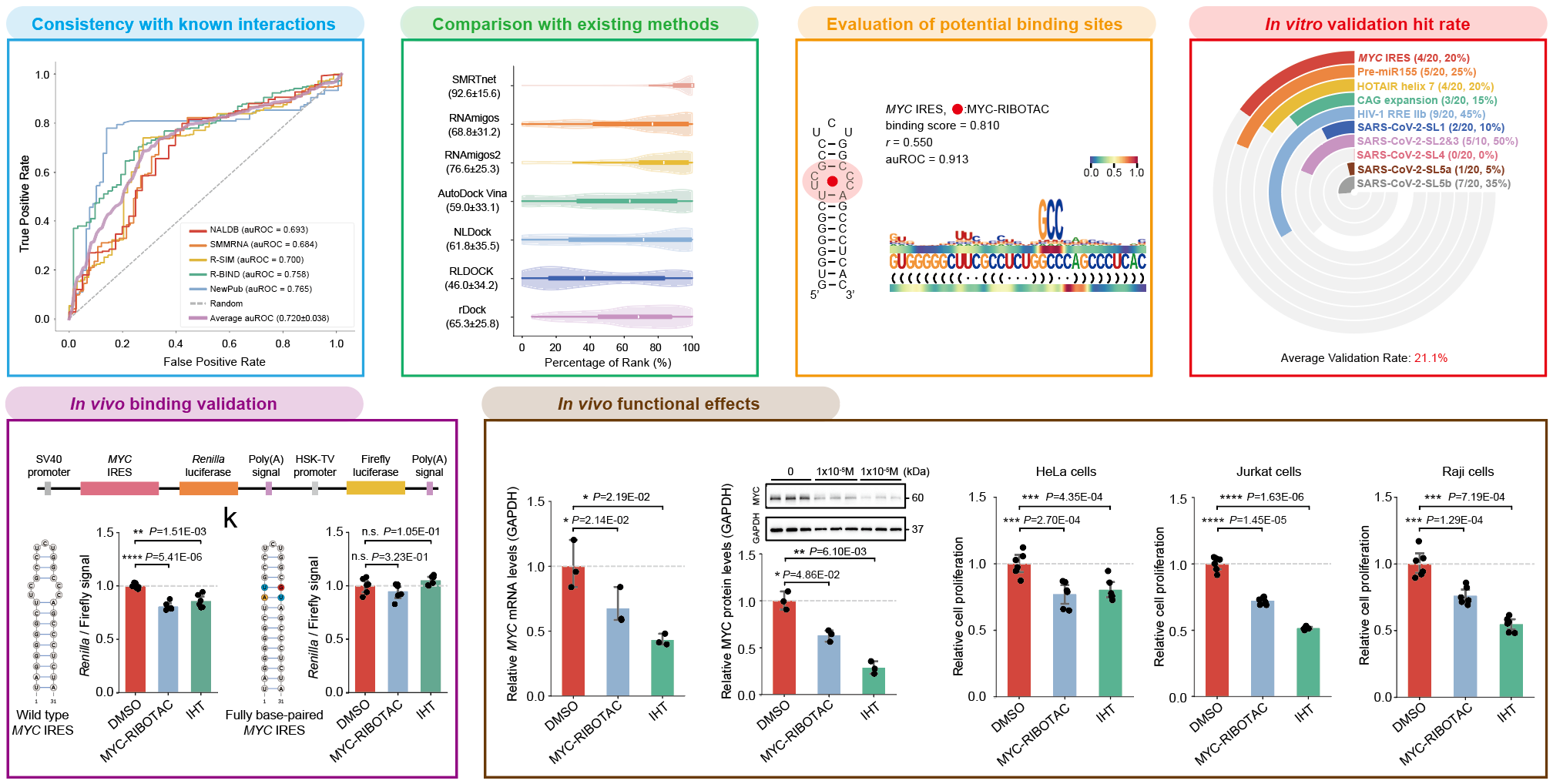
Using the cancer target MYC, which has long been considered “undruggable,” as a representative example, the research team redirected their focus to its RNA, MYC RNA, with particular emphasis on the internal ribosome entry site (IRES) element that is critical for regulating MYC expression. By integrating SMRTnet predictions with experimental validation, they identified multiple compounds capable of binding to the MYC IRES. Notably, one candidate compound markedly reduced MYC expression at both the transcriptional and translational levels and effectively inhibited cell proliferation while inducing apoptosis across three cancer cell lines, demonstrating promising antitumor activity.
In summary, the study reports the development of the artificial intelligence method SMRTnet and systematically demonstrates—through large-scale virtual screening combined with in vitro validation—its superior performance in predicting small molecule–RNA interactions and identifying binding sites. This approach substantially enhances the efficiency of early-stage drug screening and establishes a solid foundation for subsequent lead optimization and in vivo evaluation. More broadly, the work provides an efficient and reusable AI framework for the early discovery of RNA-targeted small molecule drugs, highlighting the potential of AI to accelerate the initial stages of RNA-targeted drug development.
Postdoctoral researcher Yuhan Fei from the School of Life Sciences at Tsinghua University (currently Professor at the School of Pharmacy, China Pharmaceutical University), PhD student Pengfei Wang and Jiasheng Zhang from the School of Life Sciences, Tsinghua University, are listed as the co–first authors of paper. Associate Professor Qiangfeng Cliff Zhang from the School of Life Sciences, Tsinghua University and the Beijing Frontier Research Center for Biological Structures, and Professor Yangming Wang from the School of Future Technology, Peking University, are the co–corresponding authors. PhD students Xinyue Shan, Zilin Cai, and Jianbo Ma from the School of Life Sciences at Tsinghua University also made significant contributions to this work. This work has been supported by the National Key Research and Development Project of China, the National Natural Science Foundation of China, the Beijing Natural Science Foundation, the Postdoctoral Foundation of Tsinghua-Peking Center for Life Sciences, the Beijing Advanced Innovation Center for Structural Biology, the Tsinghua-Peking Joint Center for Life Sciences, the Beijing Advanced Center of RNA Biology (BEACON), and the New Cornerstone Science Foundation through the XPLORER PRIZE.
Code link:https://github.com/Yuhan-Fei/SMRTnet
Paper link: https://www.nature.com/articles/s41587-025-02942-z
