Epigenetic information, particularly heterochromatin modifications, can be stably transmitted across cell generations, thereby maintaining cell fate stability. This phenomenon is known as "epigenetic memory." During the transition from mammalian gametes to early embryos, cell fate undergoes drastic resetting, accompanied by extensive epigenetic reprogramming. Although the reprogramming landscapes have been gradually mapped, the identification of key regulatory factors in the process and their mechanisms remain unresolved in the field.
On October 20, 2025, the research group led by Prof. Wei Xie from the School of Life Sciences at Tsinghua University published a research paper entitled "EZHIP restricts noncanonical PRC2 binding and regulates H3K27me3 intergenerational inheritance and reprogramming" in Cell Stem Cell. This study identified EZHIP (EZH inhibitory protein) as a key epigenetic reprogramming factor in mouse early embryos. It controls multi-layer H3K27me3 intergenerational reprogramming, including genome-wide H3K27me3 erasure and re-establishment, as well as the transmission of noncanonical imprinting memory.

H3K27me3 is an important repressive histone modification catalyzed by PRC2 (Polycomb Repressive Complex 2). During intergenerational reprogramming in mice, H3K27me3 exhibits highly dynamic distribution. In oocytes, H3K27me3 is distributed at canonical developmental gene promoter regions (referred to as canonical Polycomb target regions) and also appears in intergenic regions, often spanning over hundreds of kilobases (referred to as noncanonical H3K27me3 domains). After fertilization, canonical H3K27me3 at developmental gene promoters is rapidly erased and is not re-established until around implantation. However, the noncanonical H3K27me3 domains are retained after fertilization and are transiently maintained during preimplantation development [1]. Interestingly, as noncanonical H3K27me3 domains exist only on the maternal allele originated from the oocyte, the early embryo exhibits a form of genomic imprinting controlled by H3K27me3 (rather than DNA methylation), known as noncanonical genomic imprinting [2]. Why is H3K27me3 reset in some regions during intergenerational reprogramming, while it is inherited in others? What are the underlying regulatory factors? These questions remain unanswered mysteries.
To address these questions, the research team first mapped the chromatin binding of PRC2 during oocyte and early embryonic development. The authors found that in growing oocytes, PRC2 binds to both canonical Polycomb target regions and noncanonical H3K27me3 domains, actively catalyzing H3K27me3 modification. However, in fully grown oocytes (FGOs), PRC2 dissociates from chromatin. After fertilization, PRC2 reappears on chromatin during the late one-cell stage and binds specifically to the noncanonical H3K27me3 domains in an allele-specific manner. It is not until the blastocyst stage that PRC2 relocates to canonical Polycomb target regions to initiate the re-establishment of H3K27me3.
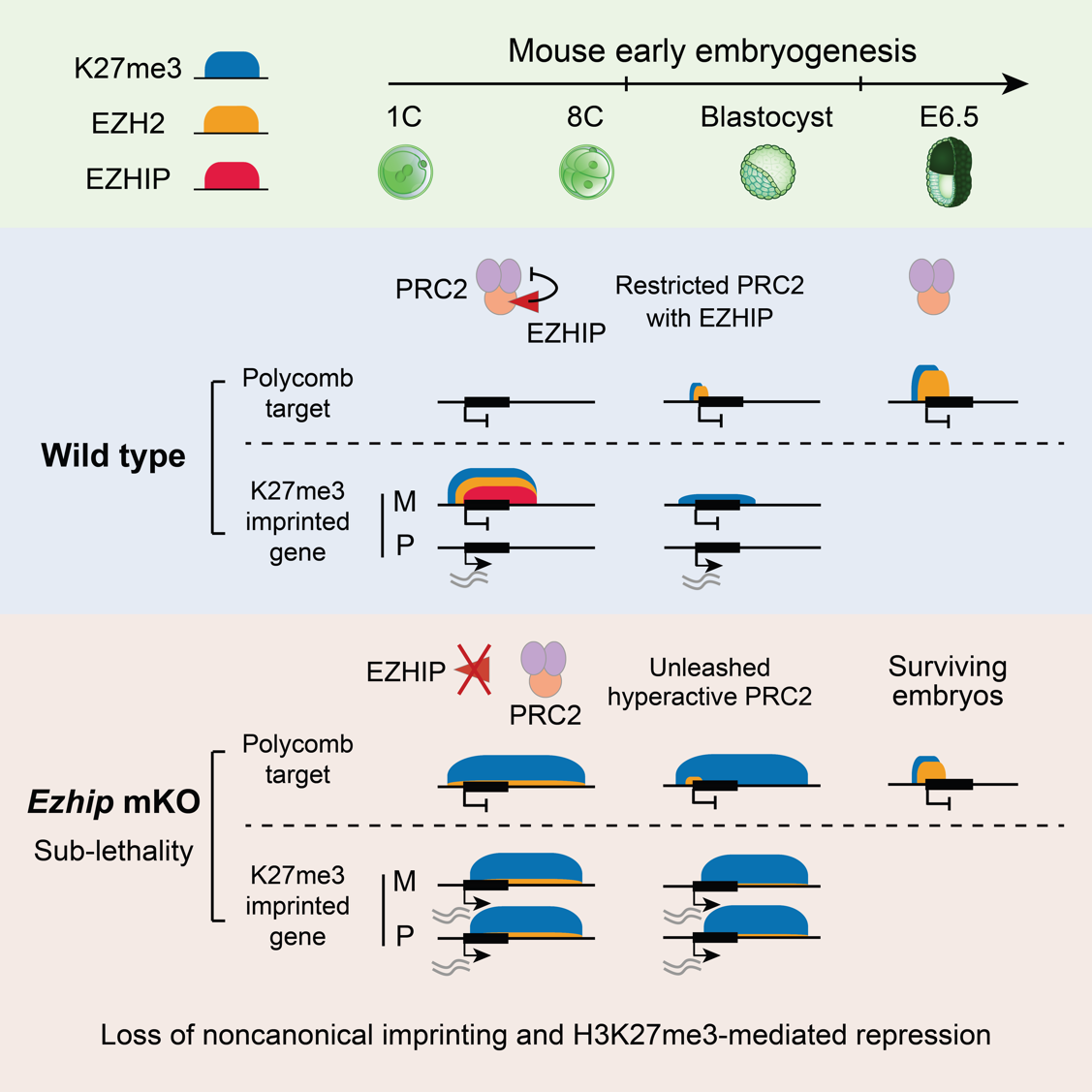
Figure 1. EZHIP regulates noncanonical PRC2 binding and intergenerational H3K27me3 reprogramming
The research team further demonstrated that EZHIP regulates PRC2 noncanonical chromatin binding and H3K27me3 intergenerational reprogramming in early embryos. EZHIP is an endogenous PRC2 inhibitor, specifically expressed in germ cells but silenced by DNA methylation in other cell types [3]. Interestingly, the authors found that Ezhip mRNA accumulates substantially in oocytes but is translationally repressed. Ezhip is actively translated only around two days post-fertilization, resulting in highly abundant EZHIP protein in preimplantation embryos. In wild-type mouse embryos, global demethylation of H3K27me3 occurs before implantation, whereas in EZHIP deficient embryos, this demethylation process is impeded, and H3K27me3 levels rise rapidly. More intriguingly, while the parental asymmetry of H3K27me3 is maintained until the blastocyst stage in wild-type embryos and mediates non-canonical imprinting memory, this asymmetry is completely lost by the two-cell stage in EZHIP deficient embryos, leading to the loss of noncanonical genomic imprinting. Further mechanistic studies revealed that in preimplantation embryos, EZHIP co-binds with PRC2 in noncanonical H3K27me3 domains, forming an H3K27me3-PRC2-EZHIP complex on chromatin, thereby restricting PRC2 enzymatic activity and spreading [4]. Upon EZHIP loss, PRC2 is no longer restricted and subsequently catalyzes H3K27me3 across the majority of the genome, masking almost all parental H3K27me3 memory. Paradoxically, this widespread H3K27me3 deposition does not cause global gene silencing, but instead results in H3K27me3 to lose its repressive function, leading to aberrant activation of noncanonical imprinted genes and defective imprinted X-chromosome inactivation. The authors hypothesized that this might be because H3K27me3 effectors (such as PRC1 and PRC2) are diluted by the massively increased H3K27me3, preventing their concentration on correct target gene regions, thus failing to mediate gene repression. Furthermore, the excessive accumulation of H3K27me3 caused by EZHIP deficiency also impedes the subsequent re-establishment of H3K27me3 at canonical Polycomb target regions and is associated with partial lethality post-implantation.
In summary, this study reveals EZHIP as a key epigenetic reprogramming factor in early embryos, regulating the erasure, inheritance, and re-establishment of intergenerational H3K27me3 reprogramming. Furthermore, these findings uncover a fundamental principle of epigenetic memory transmission: either deficient or excessive activity of heterochromatic enzymes can lead to the loss of epigenetic memory and gene repression function.
Professor Wei Xie from the School of Life Sciences at Tsinghua University is the corresponding author of this paper. PhD students Yitian Zeng and Feng Kong from Tsinghua School of Life Sciences are co-first authors. Undergraduate Zihan Xu, postdoctoral fellow Xukun Lu, PhD student Qian Li, postdoctoral fellow Bofeng Liu, intern Shu Liu, research assistants Lijun Dong, Ling Liu, and Wenying Wang at Tsinghua University also made important contributions to this project. Professor Bing Zhu from the Institute of Biophysics, Chinese Academy of Sciences, provided important guidance for this study. The project received assistance and support from the Tsinghua University Laboratory Animal Research Center. The research was funded by the National Natural Science Foundation of China, the National Key R&D Program of China, the Tsinghua-Peking Center for Life Sciences, and the China Postdoctoral Science Foundation. Professor Wei Xie is an HHMI International Research Scholar and a New Cornerstone Investigator.
Reference
1. Zheng, H., Huang, B., Zhang, B., Xiang, Y., Du, Z., Xu, Q., Li, Y., Wang, Q., Ma, J., and Peng, X. (2016). Resetting epigenetic memory by reprogramming of histone modifications in mammals. Molecular cell 63, 1066–1079.
2. Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424.
3. Piunti, A., and Shilatifard, A. (2021). The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol 22, 326–345. 10.1038/s41580-021-00341-1.
4. Jain, S.U., Rashoff, A.Q., Krabbenhoft, S.D., Hoelper, D., Do, T.J., Gibson, T.J., Lundgren, S.M., Bondra, E.R., Deshmukh, S., and Harutyunyan, A.S. (2020). H3 K27M and EZHIP impede H3K27-methylation spreading by inhibiting allosterically stimulated PRC2. Molecular Cell 80, 726–735. e727.
