DNA methylation (DNAme) plays important roles in transcriptional regulation, genomic imprinting, X chromosome inactivation, cell fate, and state transition. Several studies have revealed global reprogramming of DNAme along mammalian embryogenesis. However, a systemically integrated, technology-neutral, high resolution, cross-species analysis of DNAme-RNA expression networks during peri-implantation embryogenesis has not been revealed.
On June 3, 2025, Professor Fan Zhou's research group from the School of Life Sciences, Tsinghua University published a research article titled “Dissecting pre- to post-implantation transition of DNA methylome-transcriptome dynamics in early mammalian development” in Cell Reports. The authors revealed that major peri-implantation lineages undergo a stepwise genomic silencing with de novo DNAme. A small group of genes revealed a non-canonical active expression along with their promoter methylation during PPT. Further manipulation of DNAme indicated a complex interplay between DNAme and transcription circuitry during peri-implantation embryogenesis.
Using multi-omics sequencing, the research group selected single cells from mouse embryos spanning E3.5 to E5.5 for DNA methylation (DNAme) profiling. The results revealed a dramatic global increase in DNAme across all lineages after implantation, with the embryonic lineage—epiblast (EPI)—showing both faster and higher levels of de novo methylation compared to extraembryonic lineages, visceral endoderm (VE) and extraembryonic ectoderm (ExE). Across mouse, monkey, and human embryos, a wave of global demethylation that reaches its lowest point around implantation, after which lineage-specific DNAme patterns are re-established. Moreover, subpopulations of VE exhibit distinct DNAme profiles across genomic elements and at genes specifically expressed in each VE subtype, indicating that VE subtypes with the segregated lineage fates possibly underwent their own DNAme dynamics (Figure 1).

Figure 1. Emergence of lineage-specific DNAme in early mammalian development
Differential DNAme between parental genomes is known to regulate imprinted-gene expression and to enforce asymmetric silencing of transposable elements, yet how this parental bias evolves throughout the pre- to post-implantation transition (PPT) in mammals has not been systematically charted. Here the research group demonstrate that the erasure of parental DNAme differences follows a subtype- and element-specific timetable, and that this pattern is independent of embryo sex or mouse strain. The asynchronous dynamics observed across parental genomic compartments—particularly within transposons and retrotransposons—raise the possibility that these stage-dependent changes contribute to early embryonic gene regulation and influence lineage fate decisions (Figure 2).
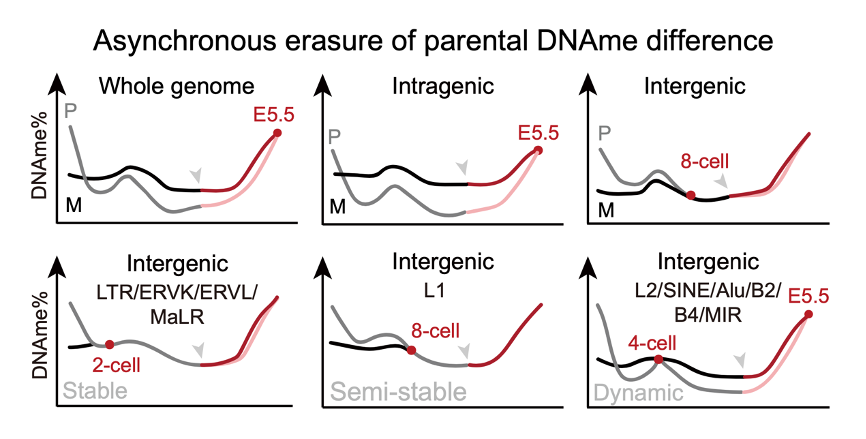
Figure 2. Asynchronous erasure of parental DNAme differences
Analysis of differentially methylated regions (DMRs) revealed a set of EPI-specific hypomethylated promoters at E5.5 whose associated genes are significantly enriched in key developmental pathways active at E6.5, most notably gastrulation. The canonical gastrulation-related gene Brachyury (T) exemplifies this pattern: its promoter acquires an EPI-specific hypomethylated state by E5.5 and is subsequently activated specifically within the primitive streak (PS) at E6.5. In contrast, VE-specific or ExE-specific hypomethylated promoters are linked to genes enriched digestive-system morphogenesis and placental development. Collectively, these lineage-specific hypomethylated promoters appear to pre-configure gene regulatory switches through an “epigenetic pre-programming” mechanism, thereby constraining differentiation toward non-target lineages and ensuring faithful lineage segregation (Figure 3).
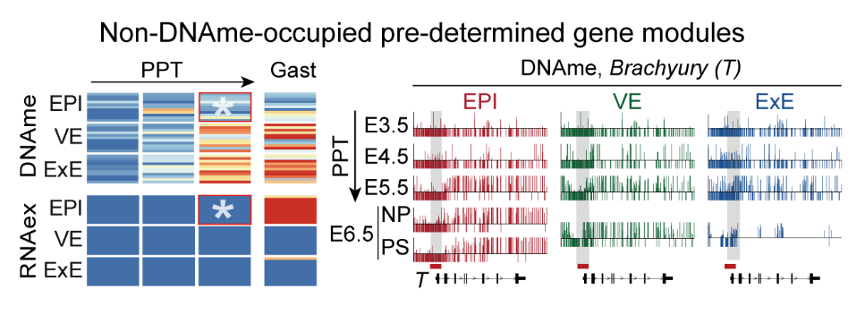
Figure 3. Establishment of non-DNAme-occupied pre-determined gene modules
Integrative analyses of DNA methylome and transcriptome reveal that, in the EPI from E3.5 to E5.5, most genes conform to the negative relationship between promoter DNAme and RNA expression (RNAex)—high promoter DNAme coincides with transcriptional repression—whereas a minority exhibit a positive, concordant coupling of promoter DNAme and RNAex. This non-canonical pattern is conserved across mouse and human. Gain- and loss-of-function experiments in Dnmt3a/b-modified mESCs, together with transcriptomic profiling of Dnmt1/3a/3b triple-knockout EPI, confirm the universality of this positive DNAme–RNA coupling in both stem-cell and embryonic contexts. Multidimensional interrogation of regulatory factors suggests that this epigenetic program is orchestrated through combinatorial mechanisms, including CTCF eviction and recruitment of lineage-specific activators such as Znf692, to fine-tune developmental gene expression during lineage specification (Figure 4).
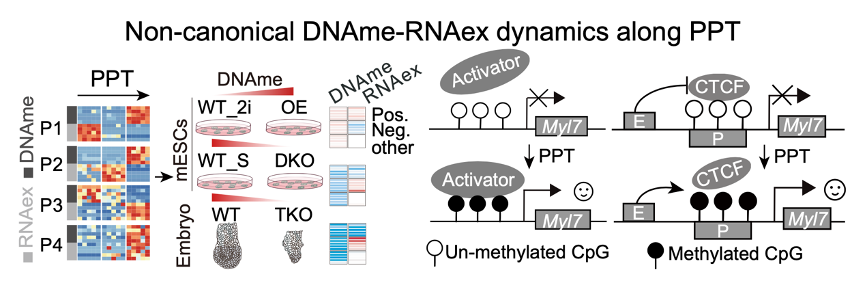
Figure 4. non-canonical DNAme–RNAex along PPT
In sum, this study uncovers lineage-specific DNAme dynamics in peri-implantation embryos and distinct DNAme patterns among VE subpopulations; it delineates the asynchronous erasure of parental DNAme differences across diverse genomic elements; and it identifies diverse promoter DNAme–RNAex dynamics during PPT, notably revealing a non-canonical positive coupling between DNAme and RNAex. These findings offer new perspectives on the interplay between epigenetic modifications and gene expression during early embryonic development.

Prof. Fan Zhou from School of Life Sciences at Tsinghua conceived the experiments and supervised the study. The co-first authors include Qingyuan Zhu (Ph.D. candidate, class of 2023) and Ying Liu (Ph.D. candidate, class of 2021). Additional contributors include Xin Hao (Ph.D. candidate, class of 2023), Shengyi Yan (Ph.D. candidate, class of 2020), Jitao Ge (Ph.D. candidate, class of 2020), and Changqing Zheng (Ph.D. candidate, class of 2023). The study was also supported by Dr. Xianwen Ren at the Changping Laboratory.
Article link: https://www.cell.com/cell-reports/fulltext/S2211-1247(25)00561-3
