Dynamic properties are essential for microtubule (MT) physiology, which depend on the polymerization and spontaneous depolymerization of tubulin isotypes at the MT ends. Mutations in tubulin-coding genes cause human diseases collectively designated as ‘tubulinopathies’. As a result, live-cell imaging of tubulins is crucial for understanding the dynamic regulation of MTs and elucidating their pathological mechanisms. Nevertheless, most existing techniques exhibit intrinsic limitations when delving into the functional specificity of tubulin isotypes and mutated tubulin variants. For instance, traditional GFP labeling fails to trace tubulins with high fidelity and efficiency, while immunofluorescence staining cannot study the dynamic behavior of MTs. Consequently, there is still a lack of unobtrusive, functional imaging of dynamic tubulin isotypes.
On Aug 19, 2024, the research group led by Guangshuo Ou published a work online in PLoS Biology entitled “AlphaFold2-guided engineering of split-GFP technology enables labeling of endogenous tubulins across species while preserving function” as a methodology article. This work employed the artificial intelligence–driven AlphaFold2 pipeline, to develop a novel strategy for the functional labeling of endogenous tubulins while preserving their inherent functionalities.
Guided by AlphaFold2, the authors implemented critical enhancements to refine the split-GFP bimolecular fluorescence complementation technique. Initially, a 16-amino acid GFP11 tag was inserted into the H1-S2 loop, a well-known flexible region of α- or β-tubulin. Subsequently, AlphaFold2-based structural prediction was utilized to optimize the linker length on both sides of GFP11 internal tag (GFP11-i), thereby facilitating stable interactions between GFP11 and the complementary fragment GFP1-10 to produce enhanced fluorescence signal (Figure 1A). Structural alignment and molecular dynamics simulation demonstrated that the intact split-GFP protein localized at the inner lumen surfaces of hollow MTs without compromising the tubulin folding process nor the overall structure of MTs (Figure 1B-C).
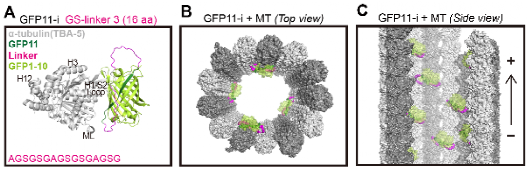
Figure 1. AlphaFold2-guided engineering of split-GFP to functionally label tubulins
To illustrate the advance of GFP11-i methodology, endogenous α- and β-tubulins were labeled with GFP11-i in C. elegans through CRISPR-Cas9 genome editing technology. Compared with traditional GFP labeling, GFP11-i resulted in less interference with multiple MT-associated biological processes such as embryogenesis or ciliogenesis. Additionally, GFP11-i methodology enabled tissue-specific labeling of endogenous tubulin isotypes and was capable of tracing the dynamics of mutated tubulin variants with high fidelity. Furthermore, to examine the applicability of GFP11-i labeling across multiple species, human α-tubulin TUBA1A (Figure 2A) and mouse α-tubulin TUBA4A (Figure 2B), all of which had been implicated in tubulinopathies, were transiently transfected into HeLa cell lines or mouse oocytes individually. Consistent with the results in worms, GFP11-i exhibits much more superior labeling efficiency and fidelity than existing technologies.
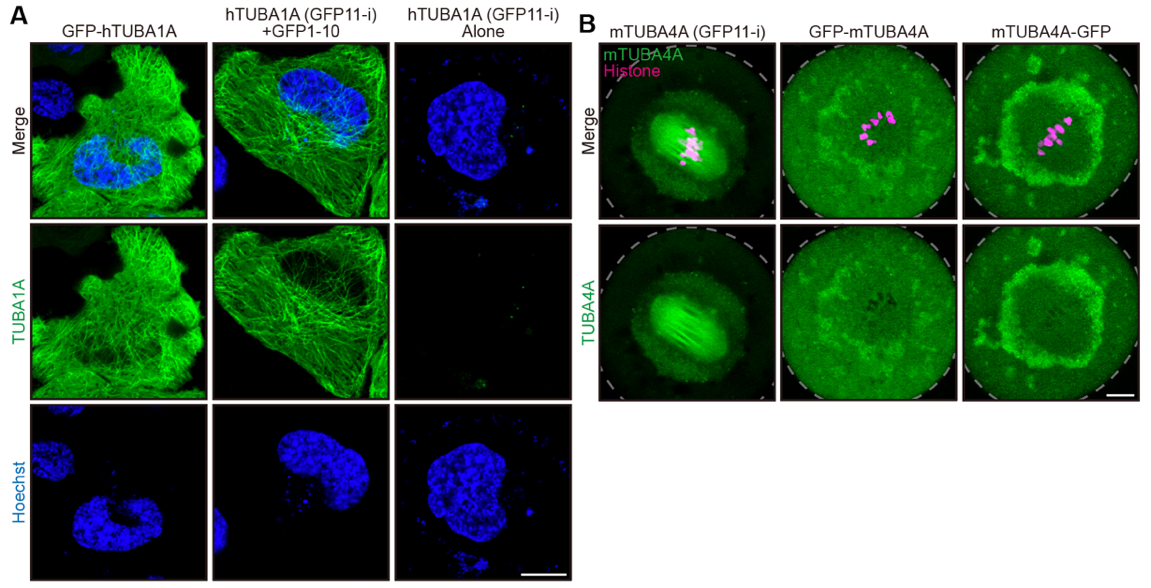
Figure 2. GFP11-i labels mammalian tubulins with high efficiency
Finally, to investigate whether GFP11-i allows for labeling of human pathogenic tubulins with high fidelity, mutated α-tubulin TUBA4A (E284G) which was implicated in human infertility was transfected into HeLa cell lines. While either untagged TUBA4A (E284G) or GFP11-i-tagged TUBA4A (E284G) disrupted cellular MT arrays (Figure 3A-C), GFP-TUBA4A (E284G) failed to induce an apparent defect, likely due to its ineffective incorporation into MT network (Figure 3A-C), suggesting the fidelity of GFP11-i labeling for live-cell imaging of tubulin isotypes in pathological contexts. In conclusion, these results underscore the considerable promise of GFP11-i methodology for understanding MT dynamics across various biological contexts, encompassing both normal physiology and tubulin mutation-associated pathologies.
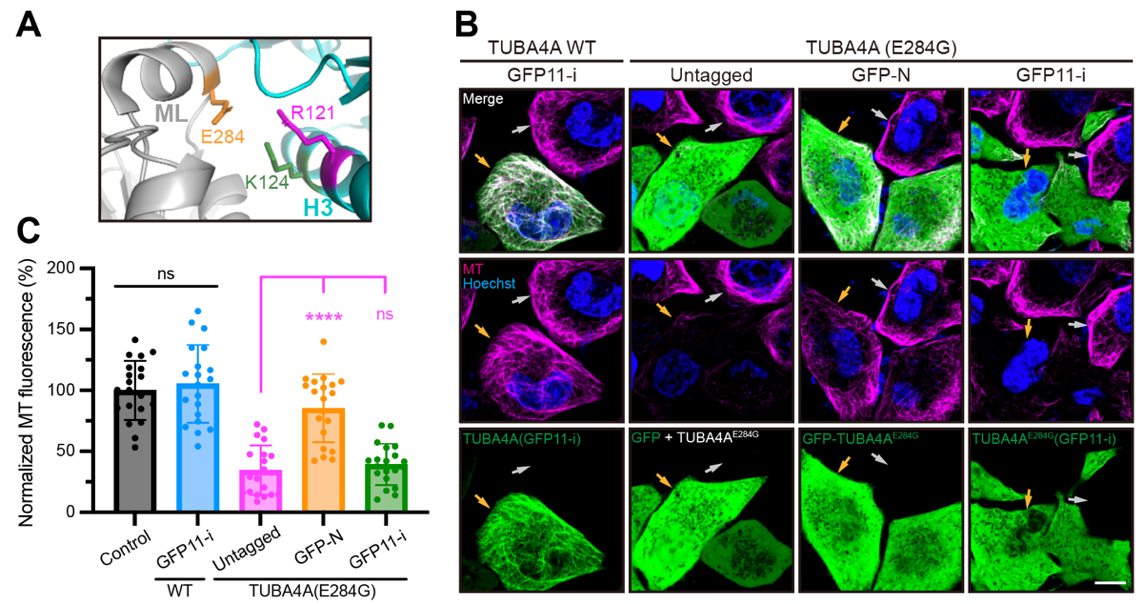
Figure 3. GFP11-i labels disease-related mutated tubulins with high fidelity
Professor Guangshuo Ou from the School of Life Sciences is the corresponding author of this article. PhD student Kaiming Xu from the School of Life Sciences is the first author of this article. All the authors come from Tsinghua University. This work was funded by the National Natural Science Foundation of China, National Key R&D Program of China, Tsinghua-Peking Center for Life Sciences, Beijing Frontier Research Center for Biological Structure and McGovern Institute for Brain Research.
A link to the paper: https://doi.org/10.1371/journal.pbio.3002615
