The maturation of mammalian oocytes and early development is a highly complex process that requires precise spatiotemporal regulation. However, the transcriptional regulatory networks and related cis-regulatory elements involved in this process are still under extensive investigation. Enhancers are critical regulatory elements that can activate and control gene expression from distal regions. Due to the limited experimental materials, the mechanisms by which enhancers regulate gene expression during mammalian oogenesis and early development remain poorly understood.
On June 5, 2024, a joint team led by Prof. Wei Xie at Tsinghua University and Prof. Jing Li at Nanjing Medical University published a research paper entitled "Mapping putative enhancers in mouse oocytes and early embryos reveals TCF3/12 as key folliculogenesis regulators" in Nature Cell Biology. This study reveals the mechanism by which active enhancers participate in transcriptional regulation during mammalian oogenesis and early development. The research provided thorough evidence supporting the existence of active enhancers in mouse oocytes, unveiled their unique features distinct from those in adult cells and tissues, and identified based on these enhancer maps key transcription factors TCF3 and TCF12 critically involved in regulating folliculogenesis.

Previous studies suggested mature oocytes and zygotes lack enhancer activities (Lawinger et al., 1999; Majumder et al., 1997). However, chromatin analyses in mouse oocytes and early embryos revealed tens of thousands of accessible regions that resemble enhancers. Researchers used STAR ChIP-seq to map a hallmark of active enhancers—histone acetylation H3K27ac — from mouse primary follicle oocytes to the inner cell mass (ICM) from blastocysts. By combining H3K27ac data from post-implantation embryos previously published from Wei Xie’s group (Xiang et al., 2020), they charted a dynamic map of putative enhancers across 16 stages from gametes to pre- and post-implantation embryos in mice. Two major transitions in the enhancer network were observed during fertilization and implantation. Active enhancers in oocytes and pre-implantation embryos were also preferentially marked by H3K4me3, correlating with low DNA methylation at these stages. Around fertilization, histone acetylation was erased genome-wide and re-established at enhancers associated with ZGA. Meanwhile, maternally inherited H3K27me3 can repress embryonic enhancers especially those near H3K27me3-controlled imprinted genes.
A large number of active enhancers in oocytes are unique and not found in early embryos or somatic cells. Therefore, the researchers further validated their activities in mouse oocytes. As a hallmark for active enhancers, bidirectional transcription was found at a fraction of enhancers in oocytes. By optimizing the high-throughput enhancer detection method STARR-seq for low-input samples, the researchers tested 70 putative enhancers with H3K27ac and bidirectional transcription, of which 64% showed positive signals. These positive enhancers were typically marked by H3K4me3 and Pol II and significantly enriched with motifs for key transcription factors in oocytes. Next, by searching for enriched TF motifs in these enhancers, the researchers identified TCF3 and TCF12 as key transcription factors in mouse oocytes. Knocking out Tcf3 and Tcf12 from oocytes led to severe defects in folliculogenesis and oocyte development, causing the transcriptome to stall at the primordial follicle stage.
In summary, the researchers identified the active enhancers from mammalian oocytes to early embryos. These results not only demonstrate the presence of active enhancers in oocytes, but also identify key transcription factors TCF3 and TCF12 that regulate folliculogenesis and oocyte development, thus shedding light on the transcriptional regulation during the oocyte-to-embryo transition.
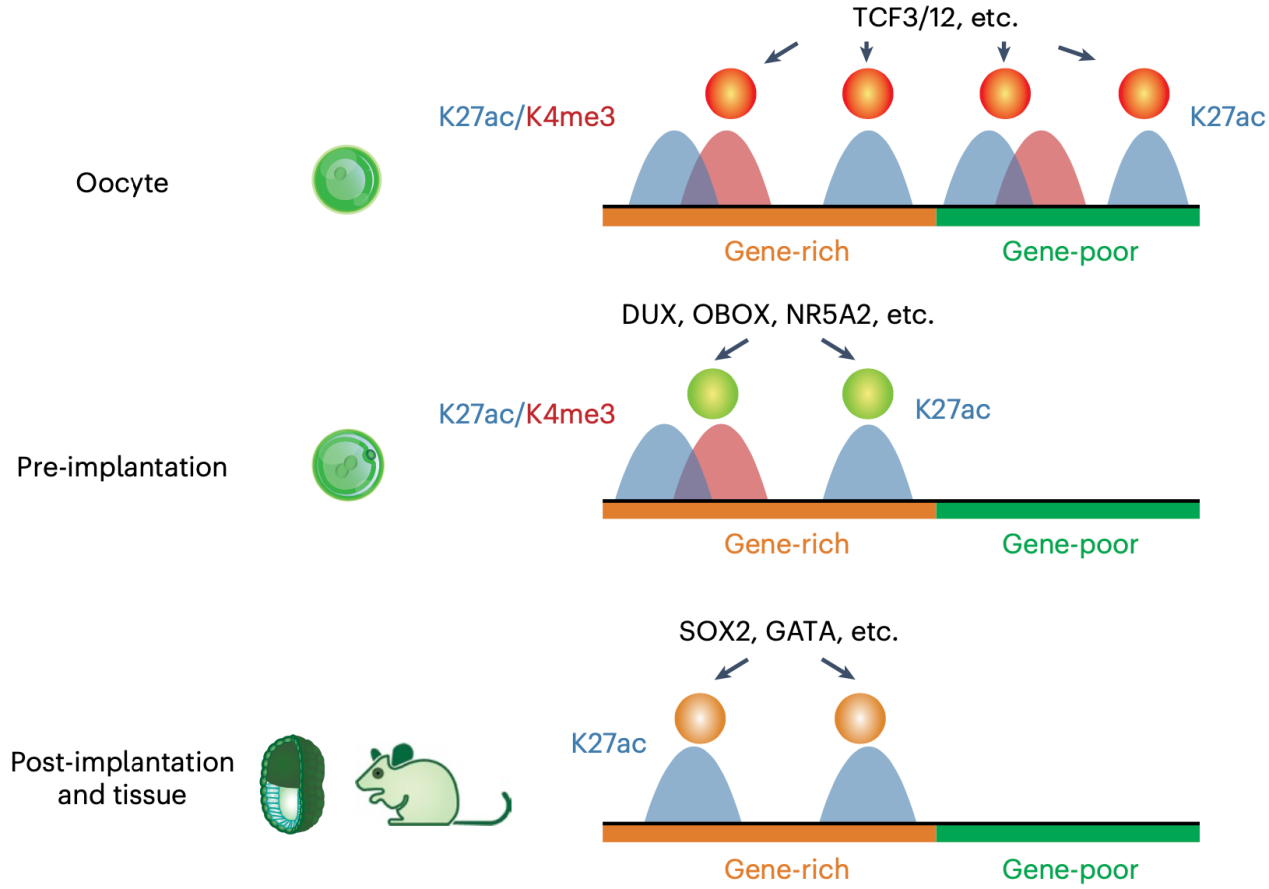
Schematic showing active enhancers in oocytes and early embryos and their interacting TFs
Prof. Wei Xie from School of Life Sciences of Tsinghua University and Prof. Jing Li from Nanjing Medical University are the corresponding authors of this work. Postdoc fellow Bofeng Liu from School of Life Sciences at Tsinghua University, Associate Prof. Yuanlin He from Nanjing Medical University, Assistant Investigator Xiaotong Wu from School of Life Sciences at Tsinghua University, Associate Prof. Zili Lin from Beijing University of Agriculture, and Ph.D. student Jing Ma from the 2019 cohort, School of Life Sciences at Tsinghua University are the co-first authors of this work. Collaborating laboratories include Prof. Jie Na’s group from School of Medicine at Tsinghua University and Prof. Maria Elena Torres-Padilla’s group from Helmholtz Zentrum München in Germany. Ph.D. student Yuexin Qiu from the 2022 cohort,Nanjing Medical University, Prof. Yunlong Xiang from Chongqing Medical University, and Ph.D. student Feng Kong from the 2022 cohort, School of Life Sciences at Tsinghua University. Prof. Wei Xie’s group at Tsinghua University also made significant contributions to this work. This study also received help from the Animal Center and the Bioinformatics core facility at Tsinghua University. Funding for this research was provided by the National Natural Science Foundation of China, the Key Research and Development Program of the Ministry of Science and Technology, and the Tsinghua-Beijing Life Science Center. Bofeng Liu is supported by postdoctoral fellowships from the Tsinghua Shuimu Scholar and the Tsinghua-Peking Center for Life Sciences. Prof. Wei Xie is an HHMI International Research Scholar and a New Cornerstone investigator.
Paper link: https://www.nature.com/articles/s41556-024-01422-x
