Natural molecular switches are ubiquitous in the biochemistry regulatory network and are critical for biological pathways. The most prominent example is the protein phosphorylation and dephosphorylation switch driven by kinases and phosphatases. In this type of switch, kinases and phosphatases perform complementary regulatory roles, leading to sophisticated up-regulation and down-regulation of diversified biochemical pathways. Taking inspiration from nature, researchers at the Molecular Architecture Design lab (MADlab) from Tsinghua University designed synthetic nucleic acid molecular switches to realize dynamic rearrangement in complicated DNA nanostructures embodying the Yin-Yang spirit.
In recent decades, DNA molecules have been found more than genetic codes, as well as functional biocompatible materials for the self-assembly of artificial structures ranging from nanometer to micrometer scales. The dynamic reconfiguration of the DNA nanostructures can be applied for various applications, such as drug delivery, molecular computing, and nanorobots. The controllable dynamics of these functional DNA nanostructures, especially those facilitated by enzymes, offer versatile gears in the toolbox of nanotechnology.
To eliminate off-target effects caused by non-specific enzymatic reactions during the reconfiguration process of DNA nanostructures, the researchers perform fantastic designs on nucleic acid sequences. With controlled reaction conditions, the enzyme selectively processes the intended site while leaving non-working sites, thus maintaining structural integrity. After screening several DNA-modifying enzymes, the authors construct two types of molecular switches (X and Y) with Bsu DNA polymerase, large fragment (Bsu DNAP) with strand displacement activity (lacking exonuclease activity), and Nt.AlwI nicking endonuclease with a strict recognition sequence. These two molecular switches control the association and disassociation of DNA nanostructures in different forms. In type X switches, Bsu DNAP drives the dissociation of the coupled structural partners (ON to OFF), while nicking endonuclease drives the association of the decoupled partners (OFF to ON). Conversely, in type Y, the identical enzymes drive an opposite reaction cycle — Nt.AlwI was used for the dissociation of the coupled structural partners (ON to OFF), while Bsu DNAP triggers the association of the decoupled partners (OFF to ON).

Figure 1. Yin-Yang diagrams of controllable rearrangement of DNA nanostructures driven by DNA-modifying enzymes. Two types of reversible molecular switches, X (left) and Y (right), are driven by the same two enzymes but with opposite reaction flow.
To synchronize two switches, the researchers achieved a one-step cut and paste operation to present the divergent activity of the enzyme. In the one-pot reaction, multiple structures were cut into diverse pieces while pasted into new shapes. The working sites of Bsu DNAP in two switches were both designed with the 8×8 lattices to reform different digital shape lattices nanostructures. The Yin-Yang’s opposite and alternate are shown in these results.
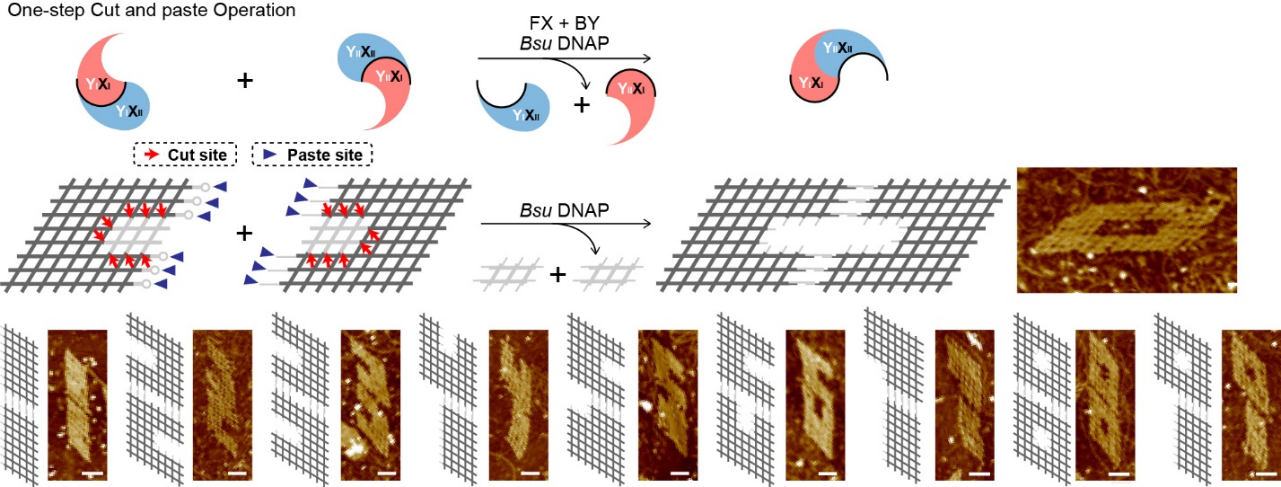
Figure 2. Controllable rearrangement of DNA nanostructures by a one-step cut and paste operation. The schematic diagram indicates that cut sites and paste sites were designed in the same lattice structure. The reaction schematic and results of the cut and paste products of digit-shaped lattices from 0 to 9 are shown.
This study is presented by the MAD lab at the School of Life Sciences, Tsinghua University. The research article, titled Synthetic molecular switches driven by DNA-modifying enzymes, was published online in Nature Communications on May 6, 2024. Dr. Hong Kang, from the School of Life Sciences, Tsinghua University (now a postdoc at the University of Pennsylvania), is the first author of this paper. Associate Professor Bryan Wei in the School of Life Sciences is the corresponding author of this paper. Associate Professor Xin Liang in the School of Life Sciences and Professor Qingshan Jia in the Department of Automation participated in the theoretical discussion of this topic.
The study was funded by the National Natural Science Foundation of China, the National Key R&D Program of China, Tsinghua University-Peking University Joint Center for Life Sciences, and other grants.
