DNA in eukaryotes wraps around the histone octamer by ~1.7 turns to form nucleosomes, the fundamental unit of chromatin. The N-termini of histone H4 interact with nearby nucleosomes, and play an important role in the formation of high order chromatin structure and heterochromatin silencing. The formation of nucleosome and heterochromatin hinders genomic DNA functions, such as replication, transcription and DNA repair. Cells have evolved mechanisms to counteract these obstacles. Acetylation of lysine residue of the histone tails neutralizes the positive charge, but also serves as molecular signals to recruit effector proteins, which regulate chromatin packaging and function in transcription activation and DNA repair.
On October 5, 2021, Zhucheng Chen’s and Xueming Li’s groups at School of Life Sciences, Tsinghua University, collaborate to publish a research article in Nature, entitled “Structure of the NuA4 acetyltransferase complex bound to the nucleosome”, which elucidates the mechanisms of nucleosome recognition and elective acetylation of H4 tail by NuA4.
NuA4 and SAGA are two important histone acetyltransferases (HAT) in yeast, which preferentially acetylate the H4 and H3 tails, respectively (Fig. 1a). NuA4 and SAGA are highly conserved, and the former is the only essential HAT for yeast survival. Recruitment of NuA4 and SAGA to promoter regions relies on the shared subunit Tra1, which interacts with different transcription factors (TFs). Because of the fundamental importance, NuA4 has been studied for more than 20 years. However, the structural basic of NuA4 assembly and the association with nucleosomes remains poorly understood.
Previously, Zhucheng Chen’s group reported the crystal structure of the catalytic subcomplex Piccolo, and a low resolution cryo-EM structure of Piccolo bound to the NCP. The group then pursues the structure of the NuA4 holo-enzyme. Due to the flexibility, it’s challenging to detect the interaction between the enzyme and the nucleosome. To reduce the flexibility, an upstream activation sequence (UAS) was engineered into the linker DNA of the nucleosome, and the TF Gal4-VP16 was added to the NuA4-NCP complex. To further restrain the flexibility, the researchers modified the histones at H4K16 to contain carboxymethyl coenzyme A (CMC), and H3 to contain trimethylated Lys36, considering that CMC stably binds to the catalytic pocket of Esa1, and the chromo domain of Eaf3 recognizes methylated H3K36. The group finally determined the structure of NuA4 bound to the nucleosome at an overall resolution of ~8.7 Å, with local resolutions of 2.7-3.4 Å (Fig. 1c).
NuA4 contains 13 subunits and is organized into two major modules: the catalytic HAT module and the transcription activator-binding TRA module (Fig. 1b). The HAT module is essentially the Piccolo subcomplex, composed of the catalytic subunit Esa1, the N-terminal fragment of Epl1, Yng2, and Eaf6. The TRA module includes the largest subunit Tra1, Actin, Arp4, Eaf2, Eaf1, and the EPc-B domain of Epl1. The two modules are linked through a disordered loop of Epl1. The structures of Eaf3, Eaf5, and Eaf7 were not resolved, and assigned to the bridge density connecting the HAT and TRA modules based on the cross-linking mass-spectrometry (Fig. 1c). The weak connection between two major modules of NuA4 explains its plasticity.
The linker DNA carrying the UAS packs against the edge of the TRA module, and projects toward Tra1 (Fig. 2a). Intriguingly, the exposed surface of Tra1 facing the UAS is highly conserved from yeast to humans, and many TF-binding sites are mapped to this region. Therefore, researchers named this surface as transcription factor binding surface (ABS). The ABS covers a large area, suggesting multiple TFs can bind the TRA module independently and/or cooperatively.
At the edge of the TRA module, a polybasic surface (PBS) formed by the Epl1 and Eaf2 subunits interacts with the linker DNA (Fig. 2b). The importance of the PBS was confirmed by the acetylation assays and the yeast genetic experiments (Fig. 2c).
The HAT module of the holoenzyme binds to the disk face of the nucleosome mainly through two elements (Fig. 3). The arginine anchors of Epl1 bind to the acidic patch of H2A-H2B, and the double function loop (DFL) of Epl1 binds to the DNA at the super-helical location 1.5. Under these conditions, the catalytic pocket of Esa1 is oriented towards the H4 N-terminus. Therefore, the modular nature of the NuA4 complex ensures that the holoenzyme employs the position-based mechanism of the catalytic Piccolo subcomplex in H4 acetylation. This is different from many histone modifiers that recognize specific sequences of the histone tails.
In summary, this work provides an integrated model of nucleosome recognition by NuA4 through the coordination of multiple elements (Fig. 1b). The PBS and ABS interact with the linker DNA, directly and indirectly, and recruit the nucleosome to the edge of the TRA module, such that the linked HAT module binds to the disk face of the nucleosome to achieve selective acetylation of the H4 tails. The relative orientation of the two major modules of NuA4 shows a large degree of plasticity, which may nevertheless provide the adaptability required in response to different transcription activators and chromatin cues. All the subunits except Eaf5 of yeast NuA4 are conserved in the TIP60 complex in human cells, suggesting that the NuA4 structure provides a good model for human TIP60.
It is important to note that the structure of the TRA module is independently confirmed by three latest studies of NuA4 (bioRxiv, extended reading).
Professor Zhucheng Chen and associate professor Xueming Li from School of Life Science, Tsinghua University, are the co-corresponding authors of this article. Graduate students Keke Qu, Kangjing Chen and Hao Wang from School of Life Science at Tsinghua University are co-first authors. This study is supported by the National Natural Science Foundation of China, Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, the National Center for Protein Science (Beijing). The cryo-electron microscope platform of Tsinghua University and the high-performance computing platform of Tsinghua University provided equipment and technical support for this research.
Original Article Link: https://www.nature.com/articles/s41586-022-05303-x
Extended reading:
https://www.researchsquare.com/article/rs-1497616/v1
https://www.biorxiv.org/content/10.1101/2022.07.11.499577v1
https://www.biorxiv.org/content/10.1101/2022.06.24.497536v1
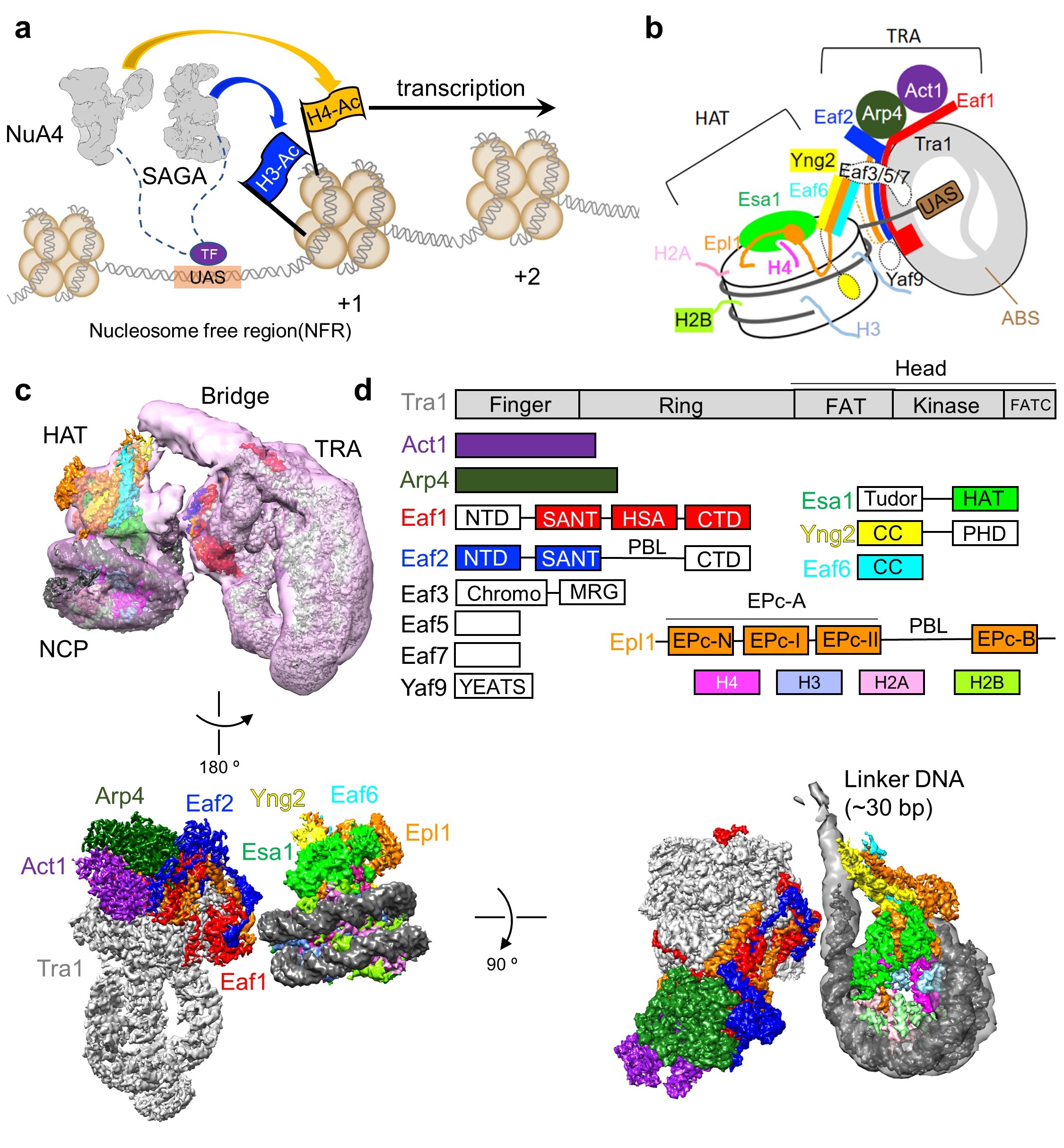
Figure 1. The working model of NuA4 and the structure of NuA4-NCP
(a) NuA4 and SAGA cooperate to regulate gene transcription. (b) Model of assembly and mechanism of nucleosome recognition of NuA4. (c) Three different views of the high-resolution composite maps of the NuA4-NCP complex. (d) Domain organization of the NuA4 subunits and histones. Proteins are color coded, with those not structurally resolved in white.
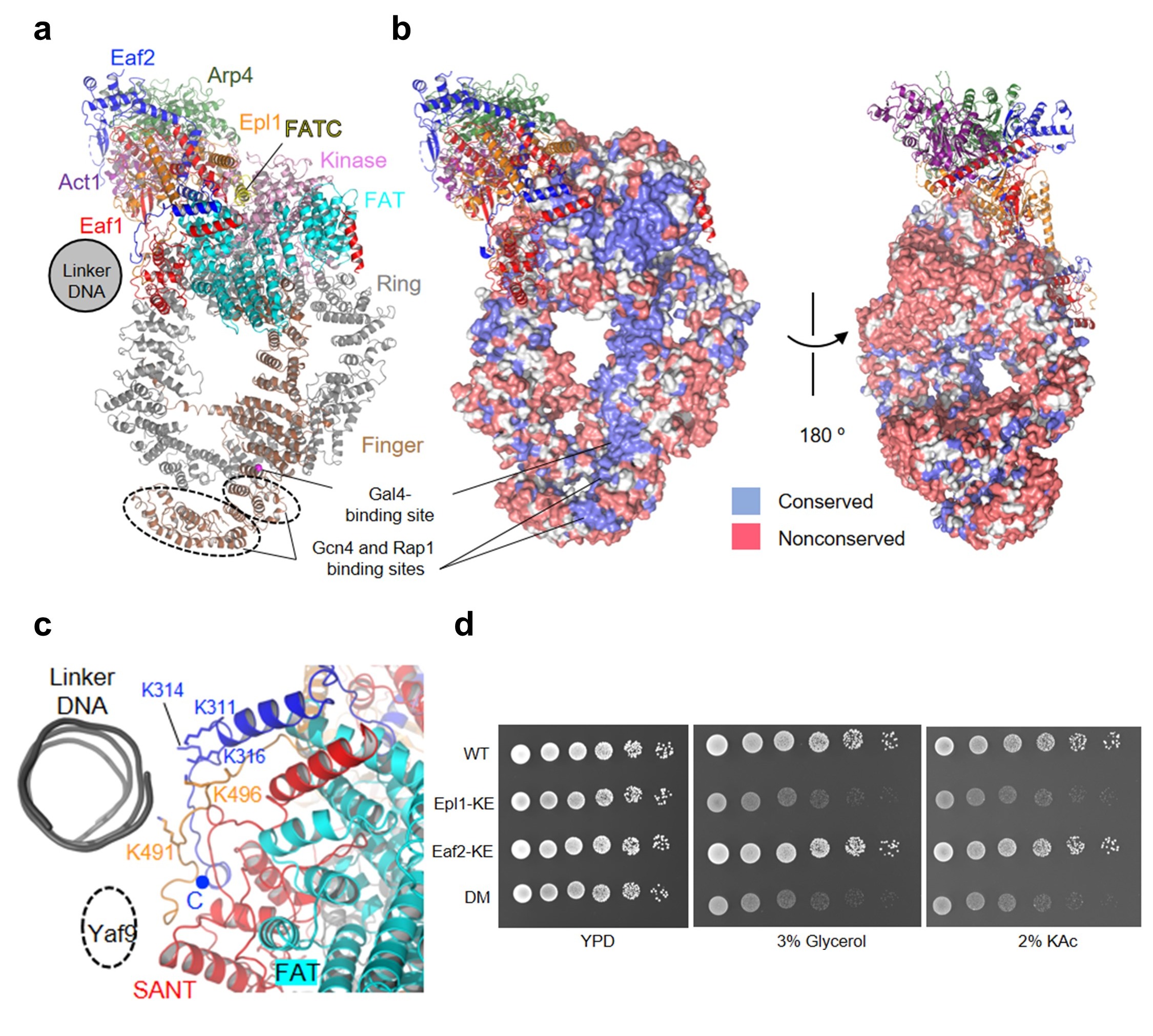
Figure 2. The TRA module of NuA4
(a) Ribbon model of the TRA module with the five Tra1 domains Ring, Finger, FAT, Kinase, and FATC colored differently. The other subunits are colored as in Fig. 1. (b) Surface conservation of Tra1. (c) Structure of the PBS region of the TRA module. (d) Growth phenotype of the WT and three mutant yeast cells under indicated conditions.
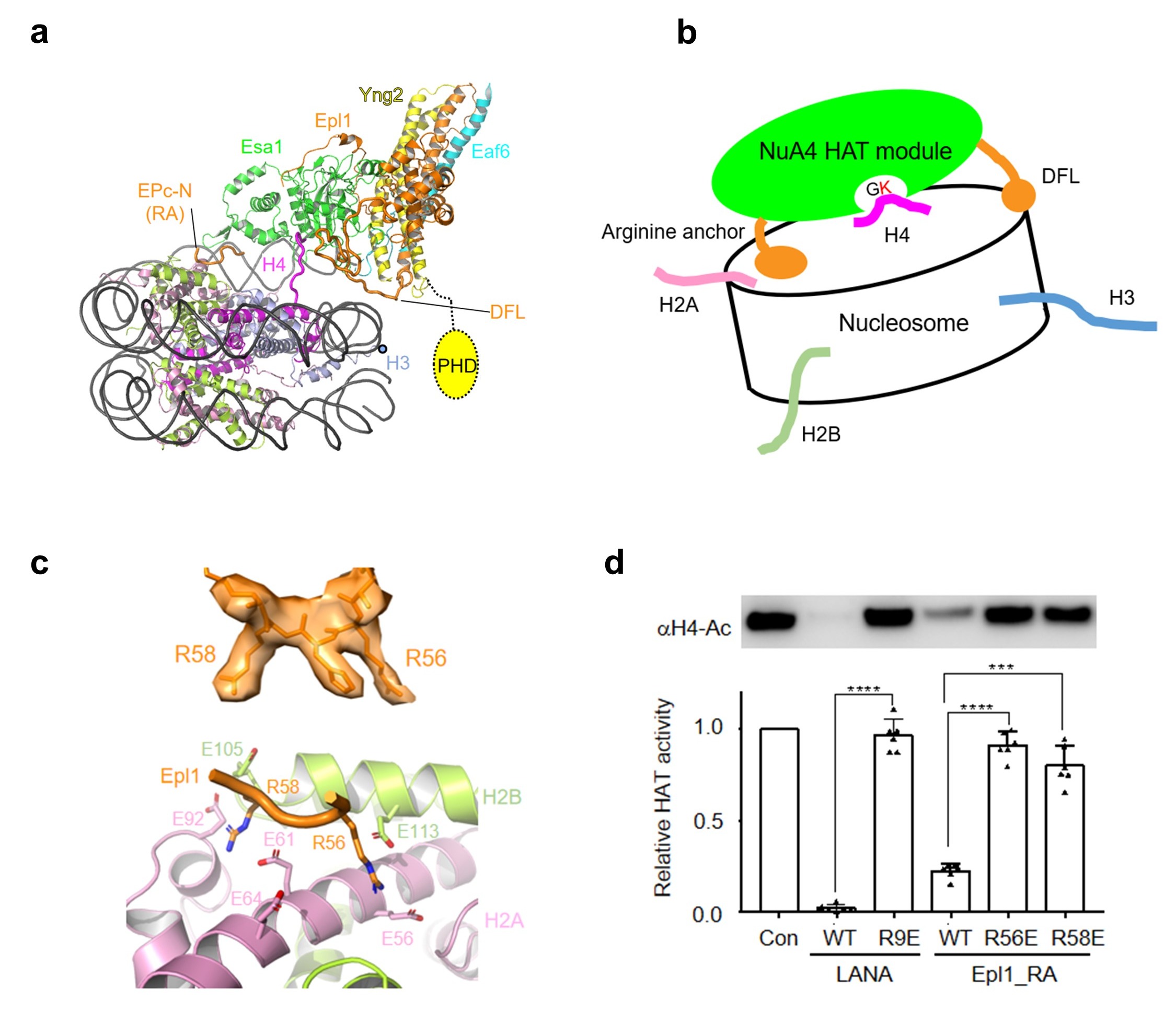
Figure 3. The mechanism of NCP recognition by the HAT module
(a) Structure of the HAT module bound to the nucleosome. RA, arginine anchor. (b) Model of the position-based mechanism of the HAT module for H4 recognition. (c) Local EM density of the arginine anchors, and the interaction with the H2A-H2B acidic patch. (d) Relative HAT activity of NuA4 to the H4 of the nucleosome in the absence and presence of the LANA and arginine anchor peptides.
