Despite the fundamental roles of TGF-β family signaling in cell fate determination in all metazoans, the mechanism by which these signals are spatially and temporally interpreted remains elusive. The cell context-dependent function of TGF-β signaling largely relies on transcriptional regulation by SMAD proteins.
On July 12, 2022, Qiaoran Xi’s research group from Tsinghua University published an article in Cell Reports entitled as “HMCES modulates the transcriptional regulation of nodal/activin and BMP signaling in mESCs”. In this study, they discover that the DNA repair-related protein, HMCES, contributes to early development by maintaining nodal/activin- or BMP signaling-regulated transcriptional network. HMCES binds with R-SMAD proteins, co-localizing at active histone marks. However, HMCES chromatin occupancy is independent on nodal/activin or BMP signaling. Mechanistically, HMCES competitively binds chromatin to limit binding by R-SMAD proteins, thereby forcing their dissociation and resulting in repression of their regulatory effects. In Xenopus laevis embryo, hmces KD causes dramatic development defects with abnormal left-right axis asymmetry along with increasing expression of lefty1. These findings reveal HMCES transcriptional regulatory function in the context of TGF-β family signaling.
Associate Professor Qiaoran XI from the School of Life Sciences, Tsinghua University is the corresponding author of this article. Tao LIANG and Jianbo BAI, are the co-first authors. Post-doc Xuechen ZHU, Dr. Hao LIN from Professor Qinghua TAO lab contributed to the Xenopus experiments. Wei ZHOU did the bioinformatic analysis, Shixin MA contributed in a part of in vitro interaction experiment.They are from Qiaoran XI's group. This work was supported by NSFC grant, MSTC grant and CLS program.
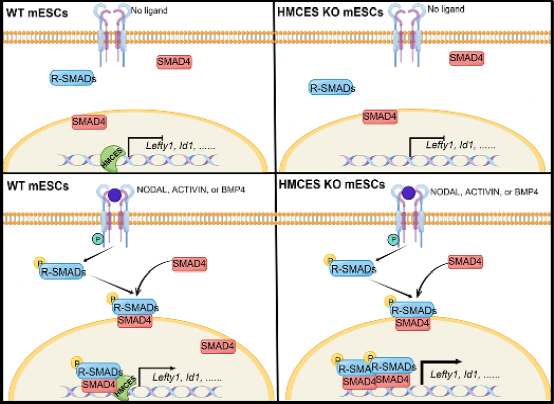
Figure 1:HMCES modulates the transcriptional regulation of nodal/activin and BMP signaling in mESCs.

Figure 2:Deletion of HMCES impairs the differentiation potential of mESCs.

Figure 3:A, Depletion of HMCES alters TGF-β family regulated transcriptome; B, HMCES interacts with SMAD; C, HMCES colocalizes with SMAD at chromatin in mESCs; D, HMCES regulates transcription through counteracting SMAD TFs association on chromatin.
原文链接:https://doi.org/10.1016/j.celrep.2022.111038
