Translation plays a key role in gene expression regulation and development. During mammalian oocyte maturation and early embryonic development, transcription is largely silenced from the full-grown oocyte until zygotic genome activation (ZGA). Therefore, during the oocyte-to-embryo transition (OET), translational regulation plays important roles in meiotic resumption, fertilization, and ZGA. However, translational studies in mammalian oocytes and pre-implantation embryos are relatively difficult due to the scarcity of experimental materials.
On June 13, 2022, Wei Xie's research group from Tsinghua University's School of Life Sciences and Lei Li's research group from Institute of Zoology at Chinese Academy of Sciences reported in the journal Nature Cell Biology entitled Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. The authors developed an ultrasensitive translatome profiling method, Ribo-lite. Using this method, the researchers investigated the translatome dynamics during OET and pre-implantation embryos, and also studied the underlying translational regulatory mechanisms. This study advanced our understanding of gene expression regulation at the translational level in early development. The ultrasensitive translatome method invented in this study also paves the way for translatome studies of other scarce samples.
Previous work has employed various mRNA-seq based translatome profiling methods, including polysome profiling (Chen et al., 2011; Potireddy et al., 2006; Sha et al., 2018; Masek et al., 2020) and Ribo-tag (Luong et al., 2020), to measure the translatome of mouse oocytes and one-cell zygotes. Ribo-seq, which added an additional step of RNase treatment to generate ribosome protected fragment (RPF), is usually more accurate than mRNA-seq based methods but requires millions of cells. Therefore, it was difficult to be adapted for studying mammalian oocytes and early embryos. In February this year, a low-input Ribo-seq method (LiRibo-seq) was developed that can accommodate as few as 5,000 cells and was employed to study the translatome of early mouse embryos (Zhang et al., 2022). In this study, by developing a highly sensitive Ribo-seq method, Ribo-lite, the authors achieved RPF-based translatome detection with only 50 HEK293 cells or single mouse oocytes. By using this technology, authors systematically investigated the translatome dynamics and regulation during mouse oocyte and early embryonic development.
This study found that the translatome and the transcriptome show distinct dynamics in oocytes and pre-ZGA embryos, but are closely coupled after ZGA. Two groups of genes, OET up-regulated and down-regulated gene, exhibit highly regulated translation. OET up-regulated genes are repressed in full-grown oocytes (FGOs) with short poly(A) tails in part through polyadenylation signal site (PAS)-proximal cytoplasmic polyadenylation element (papCPE), but are activated in MII oocytes with lengthened poly(A) tails. OET down-regulated genes (which usually have non-papCPEs or are CPE-less) are translated in FGOs with long poly(A) tails, but are translationally down-regulated in MII oocytes with shortened poly(A) tails, in part through the deadenylation complex and its adaptor protein (CNOT6L and BTG4). Many such transcripts are reactivated for translation prior to ZGA, which may participate ZGA and maternal clearance. Only those with very short poly(A) tails are destabilized.
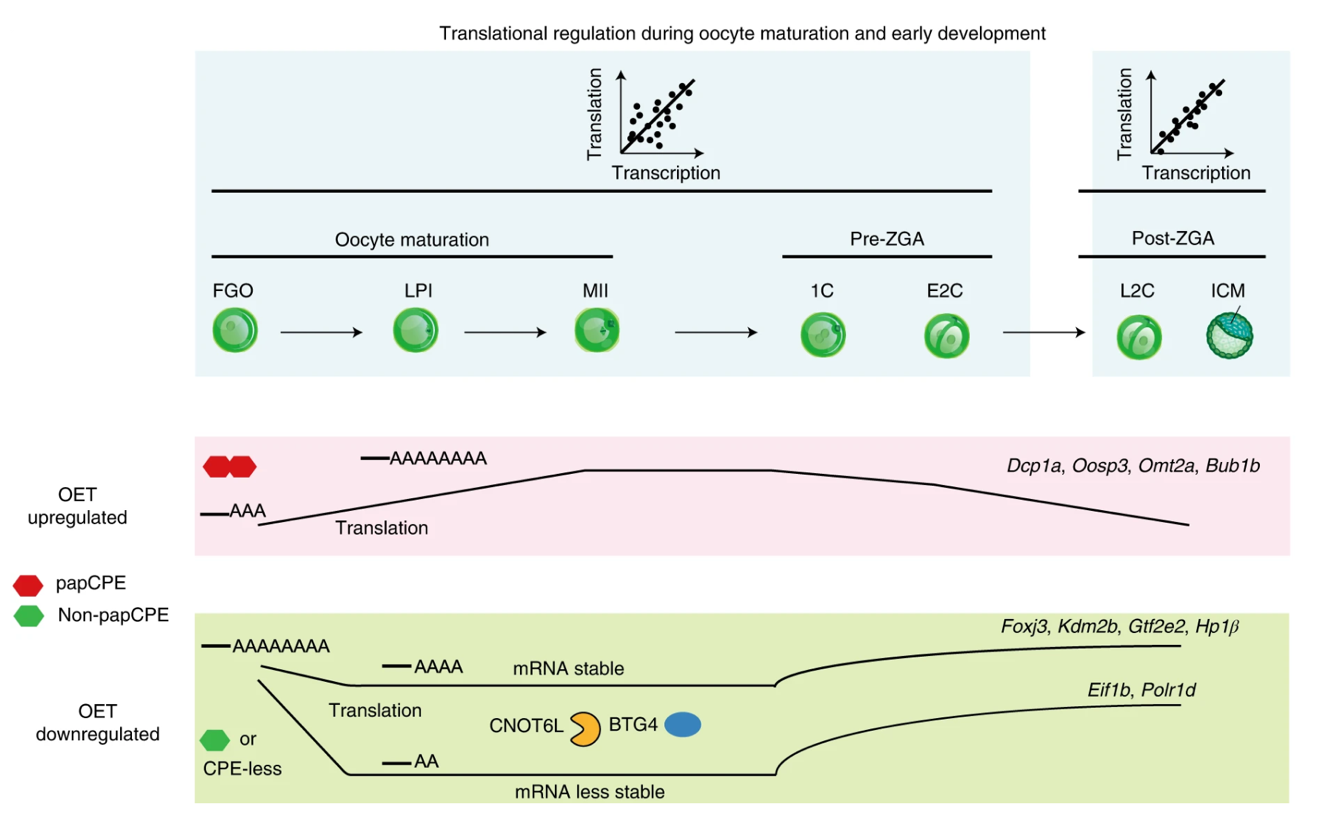
In summary, this study not only revealed highly dynamic translatome in oocytes and early embryos, but also shed light on the intimate interplay among translatome, transcriptome and poly(A) tail lengths. With the ultrasensitive Ribo-lite and the rich datasets generated, this study paves the way for further studies on translational regulation in mammalian oocytes and early development, as well as other questions that are hindered by limited materials.
Professor Wei Xie from the School of Life Sciences of Tsinghua University and Professor Lei Li from the Institute of Zoology at Chinese Academy of Sciences are the co-corresponding authors of this study. Zhuqing Xiong (a Ph.D student of the PTN program of the School of Life Sciences of Tsinghua University), Kai Xu (a Ph.D student of the CLS program of Peking University), Dr. Zili Lin (former postdoc fellow in Tsinghua University's CLS program, and currently Associate Professor at Beijing University of Agriculture), Feng Kong (research associate in Wei Xie's lab), Qiujun Wang (former postdoc fellow in Tsinghua University's CLS program), and Yujun Quan (Ph.D student in Lei Li's lab) are the co-first authors of this study. Dr. Huili Wang from the Institute of Animal Science at Jiangsu Academy of Agricultural Sciences, Dr. Haiteng Deng's research group and Xuerui Yang's research group from the School of Life Sciences of Tsinghua University, and Hengyu Fan's research group from Zhejiang University also provided essential support and help in this work. Dr. Falong Lu, Dr. Yusheng Liu and Dr. Hu Nie of the Institute of Genetics and Developmental Biology at Chinese Academy of Sciences provided advice in PAIso-seq and analysis. This project was supported by the Laboratory Animal Center and the computing core facility of the Tsinghua University. This work was funded by the National Natural Science Foundation of China, the National Key R&D Program of China, the Tsinghua-Peking Center for Life Sciences and Beijing Municipal Science and Technology Commission. Prof. Wei Xie is a recipient of an HHMI International Research Scholar award.
Link:https://www.nature.com/articles/s41556-022-00928-6
