SARS-CoV-2 is responsible for the ongoing global COVID-19 pandemic, posing serious threat to public health. The exact origin and zoonotic transmission route of the SARS-CoV-2 are still unknown. The Spike (S) glycoprotein of coronaviruses mediates viral entry by binding host receptor and fusing viral and cellular membranes. And the molecular evolution of sequence and structures of spike affect its recognition of receptors in different species. Therefore, the evolution of S glycoprotein would play important roles in cross-species transmission.
Coronavirus RaTG13, detected in the horseshoe bat Rhinolophus affinis in China’s Yunnan province, with 96.2% sequence identity to the SARS-CoV-2 genome. Pangolin coronaviruses (PCoV) closely related to SARS-CoV-2 have also been identified in smuggled Malayan pangolins (Manis javanica) in China's Guangxi (GX) and Guangdong (GD) provinces, with 85.5%-92.4% similarity to SARS-CoV-2 genome. The Professor Xinquan Wang’s group previously reported the cryo-EM structures of the RaTG13 and PCoV_GX spikes. With similar structures to SARS-CoV-2 spike, the PCoV_GX spike bound to hACE2 with an affinity comparable to the SARS-CoV-2, whereas the SARS-CoV-2 displayed high efficiency in cell entry. To explore the molecular mechanism of SARS-CoV-2 high infectivity from the point of view of S glycoprotein, Wang’s group found in the RaTG13 and PCoV_GX spikes structures, one RBD is contacted by N-glycans linked to three asparagine residues of the neighboring S protomer (N165 and N234 in the NTD and N370 in the RBD). However, the N370 glycosylation is lost in the SARS-CoV-2 S glycoprotein due to the threonine to alanine mutation at the position 372. In contrast, the typical -NST/NSS- glycosylation motif is highly conserved among 128 Sarbecovirus members. The only exception is the SARS-CoV-2 S glycoprotein with -NSA- losing the glycosylation.
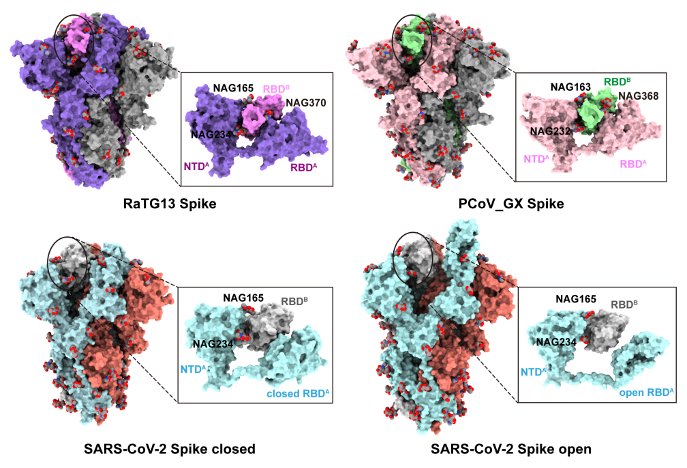
Figure 1 The unique loss of N370 glycosylation in the SARS-CoV-2 S glycoprotein.
To explore the potential roles of loss of N370 glycosylation in the SARS-CoV-2 S glycoprotein, in this study, comprehensive analysis through pseudovirus infection, cryo-EM structure determination, binding affinity measurement and molecular dynamic simulation revealed that the N370-linked glycans significant decrease the infectivity and stabilize the S glycoprotein in the “down” state, unfavorable conformation for RBD and ACE2 interaction. By removing this glycan through T732A mutation during evolution, SARS-CoV-2 adopted more desirable “up” state, thereby facilitating more efficient binding to ACE2 and more capacity to infect and transmit among humans.
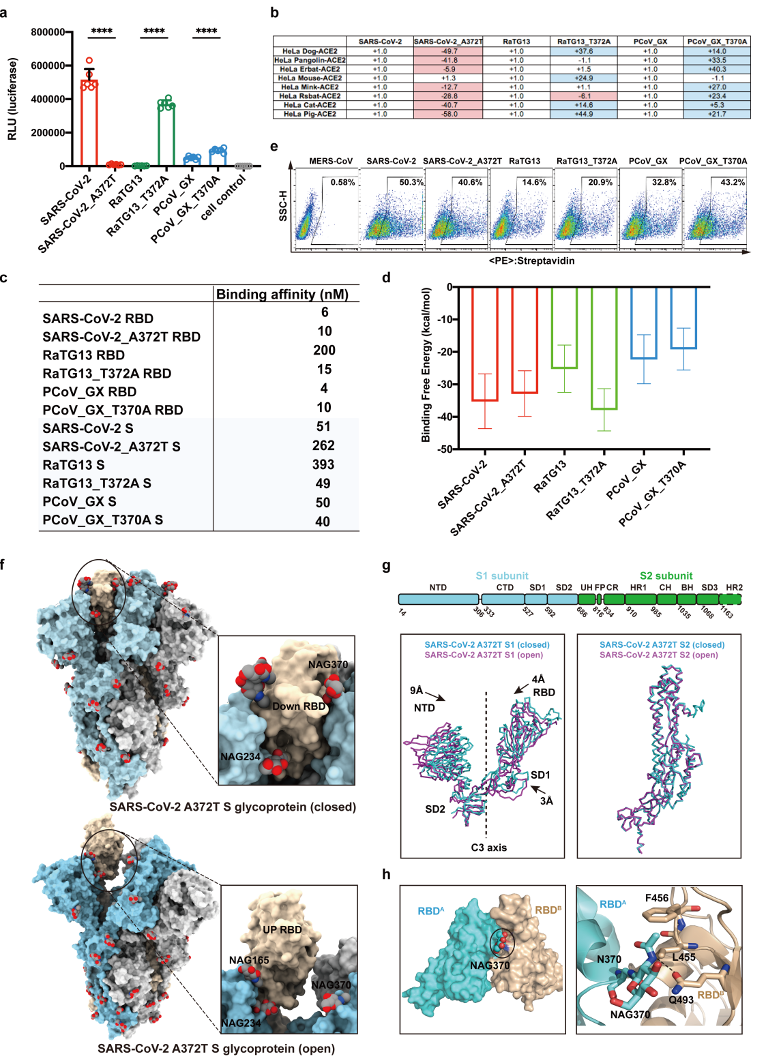
Figure 2 The effects of N370-linked glycans on the infectivity, binding to hACE2 and conformational state of the S glycoprotein.
This work was published in Cell Research on January 12, 2022, with the title of “Loss of Spike N370 glycosylation as an important evolutionary event for the improved infectivity of SARS-CoV-2”. Professor Xinquan Wang from the School of Life Sciences of Tsinghua University, Professor Linqi Zhang from the School of Medicine of Tsinghua University, and Dr. Tong Wang from Microsoft Research Asia are the co-corresponding authors. Shuyuan Zhang and Qingtai Liang are the co-first authors of this work. We thank Protein Chemistry and Proteomics Facility Technology Center, the cryo-electron microscope platform and computing platform of Tsinghua University for providing equipment and technical support for this research. This research was supported by Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, the Ministry of Education Key Laboratory of Protein Science, National Natural Science Foundation of China and Tsinghua University Spring Breeze Fund.
Original Article Link:https://www.nature.com/articles/s41422-021-00600-y
