Prof. Wei Xie’s group at School of Life Science, Tsinghua University published research in Science Advances on December 22, 2021, entitled “Methylome inheritance and enhancer dememorization reset an epigenetic gate safeguarding embryonic programs”. Their findings not only greatly deepen our fundamental understanding on how DNA methylation regulates early embryonic development in zebrafish, but also shed light on why post-fertilization global DNA methylation reprogramming is present in mammals, but is absent in non-mammalian vertebrates.
While the majority of CpG dinucleotides in vertebrate genome are hypermethylated, regulatory elements, such as enhancers and promoters, are preferentially hypomethylated. Drastic DNA methylation reprogramming after fertilization occurs to convert two terminally differentiated gametes to a totipotent embryo in mammals. However, non-mammalian vertebrates, such as zebrafish and Xenopus, inherit the global DNA methylome from gametes to early embryos. It remains puzzling why post-fertilization global DNA reprogramming occurs only in mammals but not in non-mammalian vertebrates. Despite the persisting global DNA methylation in zebrafish early embryos, previous work from Wei Xie’s group and Anming Meng’s group revealed marked local DNA methylation reprogramming at enhancers during parental-to-zygotic transition in zebrafish. Specifically, enhancers become fully methylated (thus “dememorized”), either prior to fertilization for sperm, or just after fertilization for oocyte, in zebrafish early development. Despite these intriguing findings, the function of such enhancer dememorization and how embryonic enhancers operate in a such unusually hypermethylated genome remain enigmatic.
Using a combination of powerful genetic tools and ultra-sensitive chromatin analysis methods, the researchers investigated the function of DNA methylation in early zebrafish development. By knocking down maternal dnmt1 using OMIS (Oocyte Microinjection in situ), a method previously developed to deplete maternal factors, the researchers successfully eliminated DNA methylation before zygotic genome activation (ZGA) in zebrafish, which led to embryonic lethality around gastrulation. Furthermore, these mutant embryos exhibit ectopic activation of adult enhancers and somatic genes after the loss of DNA methylation, suggesting that enhancer dememorization and global DNA methylation inheritance create an epigenetic gate that prevents premature firing of somatic enhancers and transcription programs. This, however, raises an intriguing question why embryonic enhancers are still functional despite hypermethylation. Further investigation shows the distinct DNA methylation sensitivity of embryonic and somatic enhancers is in part coded in their sequences. Somatic enhancers are preferentially CG rich, sensitive to DNA methylation and enrich CG-rich TF motifs. By contrast, embryonic enhancers are CG poor, insensitive to DNA methylation, and enrich CG-less TF motifs.
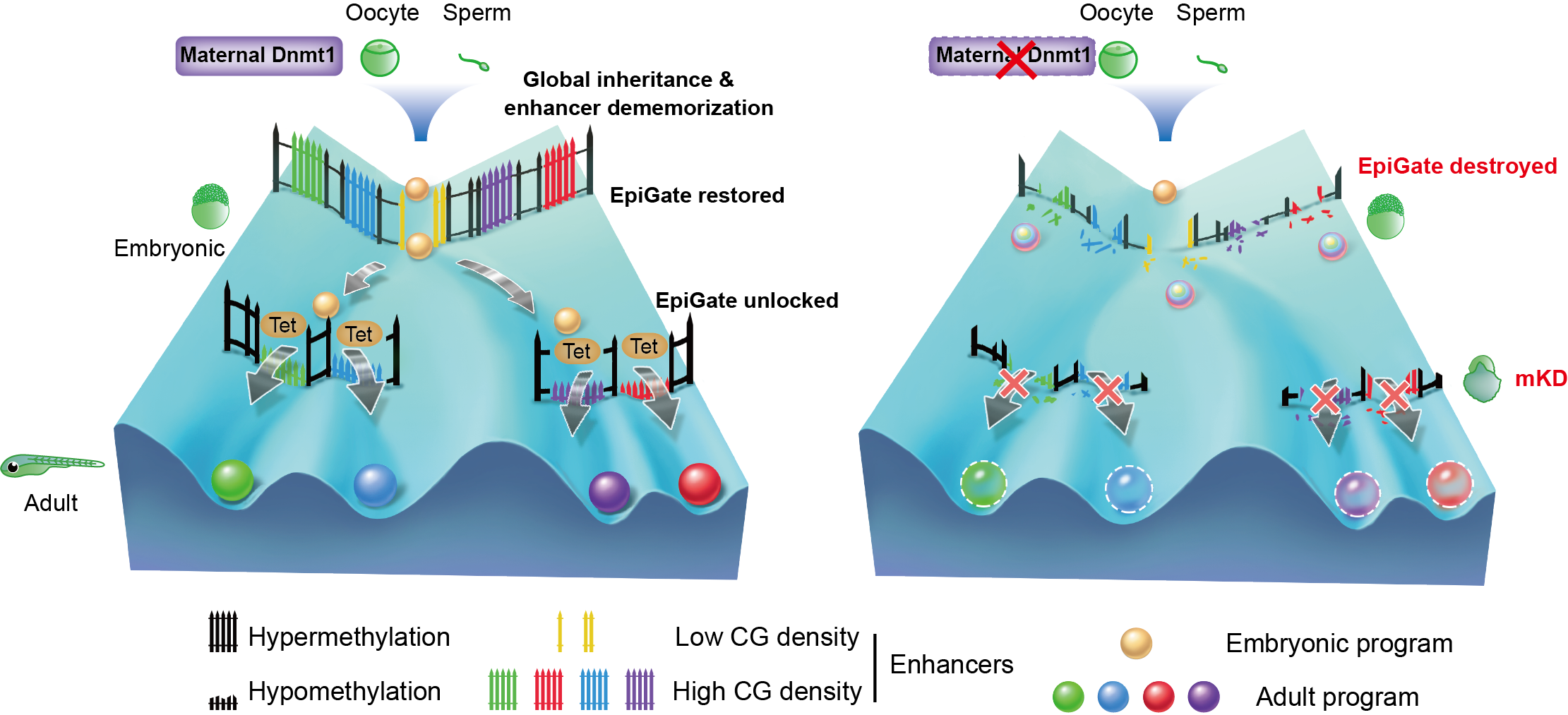
Inherited methylome coupled by enhancer dememorization resets an epigenetic gate that safeguards embryonic programs
The researchers proposed that these data may potentially unify distinct epigenetic reprogramming modes between mammals and non-mammals in early embryos to reset epigenetic memories. In this model, “dememorization” of enhancers, which resets the epigenetic clock, is achieved either through global demethylation (mammal) or global DNA methylation inheritance with local remethylation (non-mammal). Furthermore, their work on the “dememorization” of enhancers in early development may pave the way for future studies decoding how to return to totipotency during cell reprogramming and aging reversing in the future.
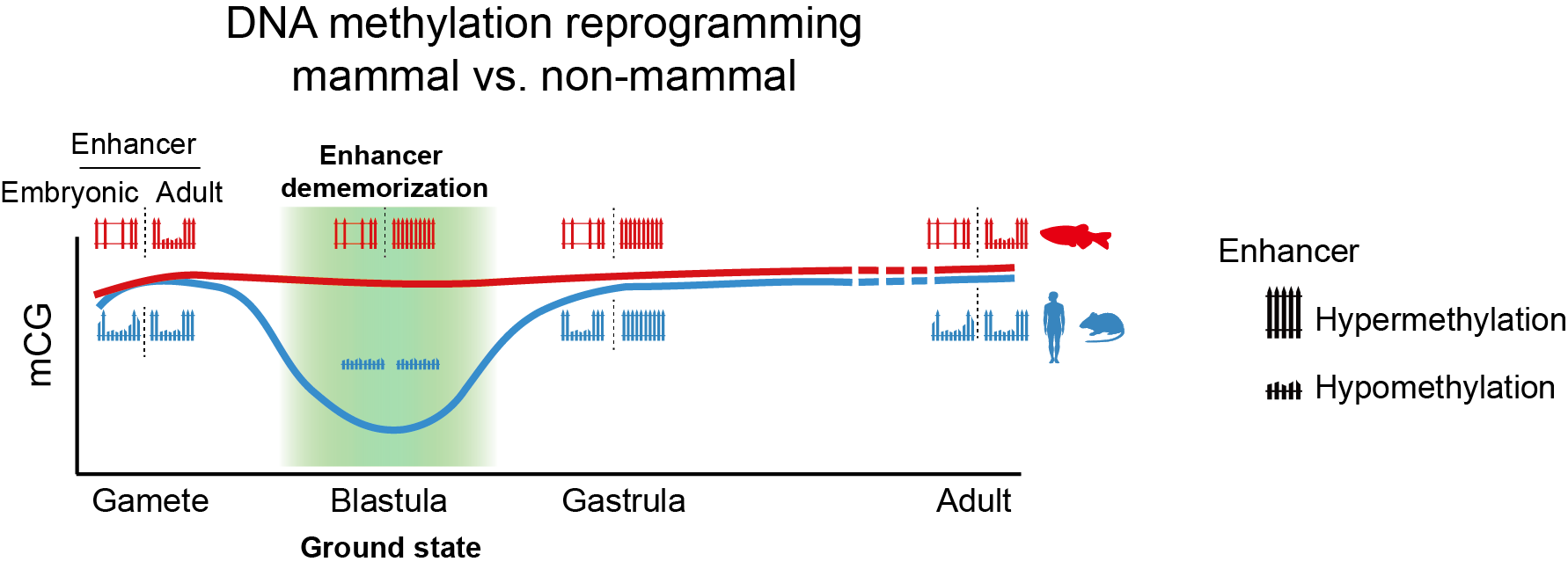
Enhancer dememorization and inherited DNA methylome reset developmental clock and unify epigenetic reprogramming modes in mammals and non-mammals
Prof. Wei Xie from School of Life Sciences of Tsinghua University is the corresponding author of this work. Postdoc fellows Xiaotong Wu and Hongmei Zhang, from School of Life Sciences at Tsinghua University, former PhD student Bingjie Zhang from the CLS program, School of Life Sciences at Tsinghua University are the co-first authors. Prof. Anming Meng from School of Life Sciences of Tsinghua University provided zebrafish facilities. Dr. Ryohei Nakamura and Prof. Hiroyuki Takeda from Graduate School of Science of University of Tokyo, Prof. Feng Liu from Institute of Zoology at Chinese Academy of Sciences also made important contributions to this work. This study was supported by the funding from National Natural Science Foundation of China, the National Key Research and Development Program of China, the THU-PKU Center for Life Sciences, Beijing Municipal Science & Technology Commission, and the Strategic Priority Research Program of the Chinese Academy of Sciences. Prof. Wei Xie is also an HHMI International Research Scholar.
Paper link: https://www.science.org/doi/10.1126/sciadv.abl3858
