A research article describing the first atomic models of human Dicer and its complexes with a pre-miRNA substrate was published online in Cell on April 26, 2018. This is a work from Hong-Wei Wang’s group in the School of Life Sciences at Tsinghua University and the Beijing Advanced Innovation Center for Structural Biology. In this article, named “Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate”, the high resolution structure of full length human Dicer (220 kDa) was determined for the first time【1】. Furthermore, this work reported two different conformers of Dicer-TRBP bound with its pre-miRNA substrate pre-let-7.
RNA interference (RNAi) pathway is a major gene regulation process in eukaryotic cells and is a widely used and powerful tool in knocking-down gene expression. In 2006, scientists Andrew Fire and Craig Mello won the Nobel prize in physiology or medicine for their discovery of the RNAi mechanism. The mostly studied endogenous small RNAs for RNAi are miRNAs. Till now, there are more than 1,800 miRNA genes discovered in human cells. Now, more and more data verified that tumors are highly related with the abnormal level of miRNAs. In human cells, most miRNAs were generated by an RNase enzyme called Dicer. It is very interesting that there is only one copy of Dicer in human cells. This means that human Dicer protein plays a pivotal role in the maturation process of most miRNAs. Dicer protein is a double stranded RNA specific RNase comprising of multi-domains. Some domains are responsible to bind RNA substrates, some domains take charge for cleaving RNA substrates and others serve as ruler for precise cleavage. Human Dicer can recognize and process variant precursor miRNAs into mature miRNAs with common features characterized by ~22 bp in length and a 3 terminal bearing 2 nt overhangs. However, there is no three-dimensional structure with high resolution of the whole human Dicer protein and the mechanism underlying Dicer’s precise RNA processing remains elusive.
The challenge in revealing the 3D structure of full-length human Dicer at high resolution lies in the following reasons: human Dicer is a 220kDa protein without symmetry, relatively small for single particle cryo-EM reconstruction; it is hard to get monodispersed human Dicer protein molecules in a homogeneous distribution in vitreous ice; human Dicer suffers from low contrast and preferential orientation in the cryo-EM specimens.
There were few 3D structures of full-length human Dicer reported at rather low resolution using singe particle EM analysis【2,3,4】. Through a decade’s effort, Hong-Wei Wang’s group conquered the technical obstacles one by one to achieve high resolution structure of human Dicer. They established biochemical procedures to purify human Dicer in complex with its cofactor protein TRBP by co-expressing the two proteins in 293F mammalian cell lines. They tried different kinds of cryo-EM grids and parameters to optimize the cryo-EM specimen preparation conditions for the molecules to distribute evenly with less preferential orientations in vitreous ice. Using an optimized image processing procedure, they were able to solve the structure of human Dicer complexed with TRBP at 4.4 Angstrom resolution by cryo-EM. In this structure, almost all the domains within human Dicer are determined and located precisely for the first time (Figure 1).
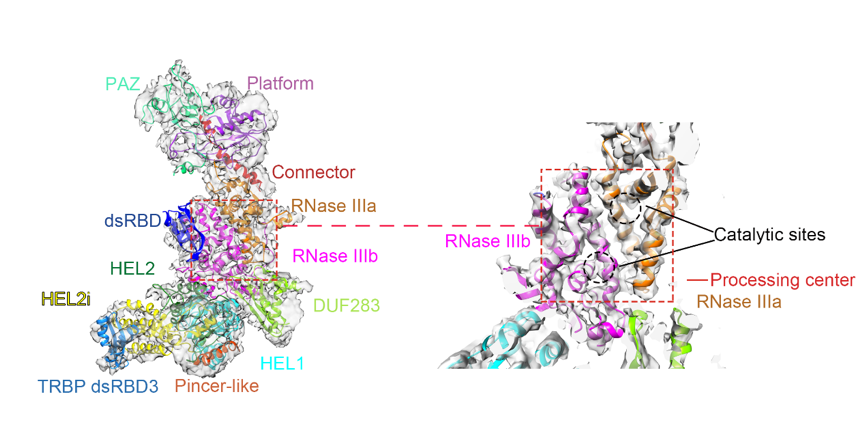
Figure 1. Cryo-EM structure of human Dicer-TRBP complex
To understand the processing of human Dicer over its RNA substrates, a complex comprising of human Dicer, TRBP and pre-let-7 RNA was assembled via in vitro reconstitution. Using cryo-EM, the Wang group also obtained the 3D reconstructions of such a complex at high enough resolution to identify two different conformers of pre-let-7 RNA present in the complex (Figure 2). In the first conformer, the stem of pre-let-7 adopts a perfect base-paired A form helix, while in the second conformer, the stem of pre-let-7 is partially splayed. In collaboration with Qiangfeng Cliff Zhang’s group at Tsinghua University with an expertise in icSHAPE technology, the research team found that the stem of pre-let-7 RNA adopts multiple conformations in free states but the loading of pre-let-7 onto human Dicer and TRBP complex may promote the stem of pre-let-7 into a stable base-paired state. These structures therefore revealed a function of the hDicer-TRBP complex in stabilizing the stem duplex of pre-let-7 in a substrate-loading state and revealed a mechanism that human Dicer uses to warrant precise dicing on pre-miRNAs with various structural features.

Figure 2. Two conformers of human Dicer-TRBP-pre-let-7 complex
Furthermore, the high resolution structure revealed that human Dicer’s DExD/H-box helicase domain presents a C-shaped architecture composed of Helicase 1, Helicase 2i and Helicase 2 subdomains. Based on sequence alignment and secondary structural analysis, the DExD/H-box helicase domain is proved to be conserved with those of RIG-I and MDA5 proteins, also involved in RNA transaction. Similarly to the latter two, human Dicer’s DExD/H-box helicase domain was proved to play important roles in regulating the dicing activity of human Dicer protein. This work opened new doors to investigate the mechanism and regulation of small RNA biogenesis in more detail in the future and may lead to the development of new RNAi tools.
Professor Hong-Wei Wang is the corresponding author of this article. Postdoctoral fellows Dr. Zhongmin Liu and Dr. Jia Wang, graduate students Hang Cheng and Ke Xin of Tsinghua University are co-first authors. Professor Qiangfeng Cliff Zhang and his graduate student Lei Sun performed the RNA sequencing and data analysis. The project was performed on the cryo-EM facility and high-performance computational facility of Tsinghua University Branch of the National Center for Protein Sciences (Beijing). This work was supported by research funds from the National Natural Science Foundation of China, the Ministry of Science and Technology, Beijing Municipal Science and Technology Commission, Tsinghua-Peking Joint Center of Life Sciences, and Beijing Advanced Innovation Center of Structural Biology.
Links: https://doi.org/10.1016/j.cell.2018.03.080.
