Higher organisms have evolved the sophisticated immune system in their long-term competition with pathogenic microorganisms. The immune systems can be divided into two parts: innate immunity and adaptive immunity. The former, which is a universal and ancient form of host defense mechanism, can be initiated through perception of pathogens by a series of germline-encoded pattern recognition receptors (PRRs). The NOD-like receptor (NLR) family is the largest group of intracellular PRRs. Perception of pathogen- or host-derived signals by NLRs results in formation of large protein complexes termed inflammasomes mediating host immune defense responses. In vivo the activity of NLR inflammasomes is stringently regulated and dysregulation of NLRs can lead to various immune-related diseases.
The NLR NLRP1 is the first member identified to form inflammasomes and its mutations are found in cutaneous immune diseases such as vitiligo. In addition to the canonical NOD, LRR and CARD domains present in NLRs, NLRP1 also possesses a unique FIIND domain (Figure 1a). Auto-cleavage of this structural domain is required for NLRP1 activation. Formation of the NLRP1 inflammasome is negatively regulated by the dipeptidyl peptidases DPP8/9. The underlying mechanism, however, remains elusive.
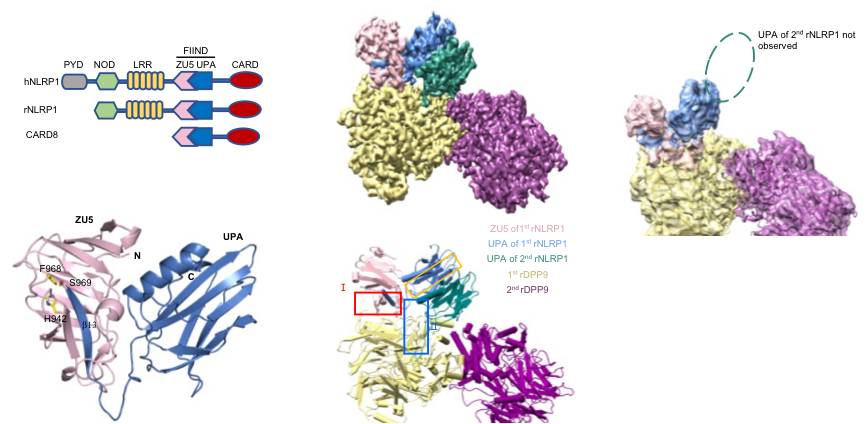
Figure 1 Crystal structure of the NLRP1 FIIND domain and Cryo-EM structure of the full-length NLRP1-DPP9 2:1 complex.
On March 17th 2021, Jijie Chai's group at Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing, China, and Franklin Zhong's group at Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, published a paper entitled “Structural and biochemical mechanisms of NLRP1 inhibition by DPP9”, elucidates the mechanism of DPP9-mediated inhibition of NLRP1 through structural biology, biochemistry and cell biology, and sheds new insights into the activation of the NLRP1 inflammasomes.
Jijie Chai's group firstly resolved a crystal structure of the rat NLRP1 FIIND domain (Figure 1b), unveiling the mechanism of NLRP1 autoinhibition mediated by the ZU5 subdomain of FIIND (Figure 1a). The group then obtained a high resolution Cryo-EM structure of the full-length rat NLRP1-DPP9 complex (Figure 1c-d). Unexpectedly, the structure revealed that rNLRP1 and rDPP9 form a 2:1 complex containing an auto-inhibited rNLRP1 molecule and an active rNLRP1 UPA-CARD fragment. Through collaboration with Franklin Zhong's group and Bin Wu's group at Nanyang Technological University, they showed that the complex functions to inhibit activation of the NLRP1 inflammasome by sequestering the active fragment and fortifying NLRP1 autoinhibition. Furthermore, they provided evidence that both NLRP1-binding and enzymatic activity are required for DPP9 to suppress NLRP1 activation in human cells. The study also implies the existence of an unidentified DPP9 substrate required for NLRP1 inhibition, and suggests that the 2:1 NLRP1-DPP9 complex may act as a bona fide receptor sensing diversified signals to mediate innate immjune responses (Figure 2).
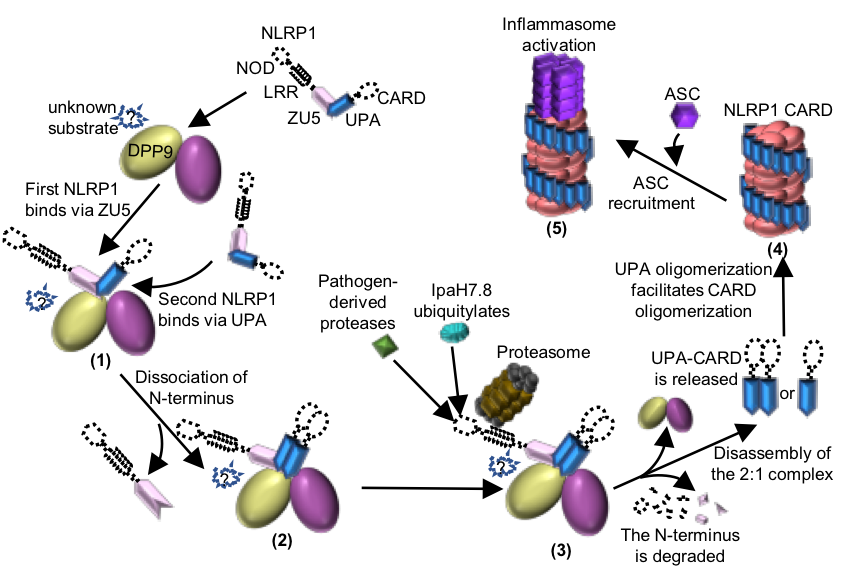
Figure 2 Working model of the DPP9-mediated inhibition of NLRP1 and the pathogen-induced activation of NLRP1.
Professor Jijie Chai from School of Life Sciences, Tsinghua University, and assistant professor Franklin Zhong from School of Medicine, Nanyang Technological University, Singapore, are the co-corresponding authors of this paper. Menghang Huang, a postdoctoral fellow in the School of Life Sciences of Tsinghua University, and Xiaoxiao Zhang, a former postdoctoral fellow in the School of Life Sciences of Tsinghua University, are the co-first authors of this article. Gee Ann Toh from Nanyang Technological University School of Medicine, assistant Professor Bin Wu and Qin Gong from Nanyang Technological University School of Biological Sciences, Dr. Zhifu Han, associate Researcher at the School of Life Sciences, Tsinghua University, and Dr. Jia Wang, assistant Researcher at the School of Life Sciences, Tsinghua University, also contributed to this research. Members of Dr. Bruno Reversade's laboratory provided help and advice for the research. The National Center for Protein Science (Beijing), the cryo-electron microscope platform of Tsinghua University, the high-performance computing platform of Tsinghua University and the Shanghai Synchrotron Radiation Light Source provided equipment and technical support for this research. This research was supported by Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking University Joint Center for Life Sciences, the National Natural Science Foundation of China (31421001 to J.C.),the Alexander von Humboldt Foundation (a Humboldt professorship to J.C.), Max Planck-Gesellschaft (a Max Planck fellowship to J.C.), Ministry of Health, Singapore, NMRC grant (MOH-000382-00 to W.B.), Concern Foundation (F.L.Z), Nanyang Assistant Professorship (F.L.Z.) and the National Research Foundation fellowship (F.L.Z.).
Original Article Link:https://www.nature.com/articles/s41586-021-03320-w
