Genome stability and integrity is essential for the survival of all organisms. However, due to the endogenous metabolites and environmental agents (such as UV light or chemical exposure), DNA damage constantly happens and posts a threat. DNA glycosylase is a DNA-binding protein responsible for repairing the DNA damage. It remains a puzzle how glycosylase can efficiently and accurately recognize DNA lesions among millions and billions of normal base pairs in the genome. It has been hypothesized that glycosylase accomplishes this task by an alternating search between two diffusion modes: a high-speed-low-accuracy mode and a low-speed-high-accuracy mode. However, due to the limitation in the spatial and temporal resolutions of current experimental techniques, the slow mode has not yet been detected.
In order to understand the molecular mechanism how glycosylase AlkD recognizes the DNA damage, Prof. CHEN Chunlai’s group from School of Life Sciences, Tsinghua University, collaborating with Prof. ZHANG Lu from Fujian Institute of Research on the Structure of Matter of Chinese Academy of Sciences, Prof. HUANG Xuhui’s group from Hong Kong University of Science and Technology and Prof. ZHAO Xinsheng’s group for Peking University, has integrated the experimental and computational methods to characterize the dynamic diffusion of glycosylase AlkD along an double-stranded DNA (dsDNA) at the molecular level. The study was published in Proceedings of the National Academy of Sciences of the United States of America.
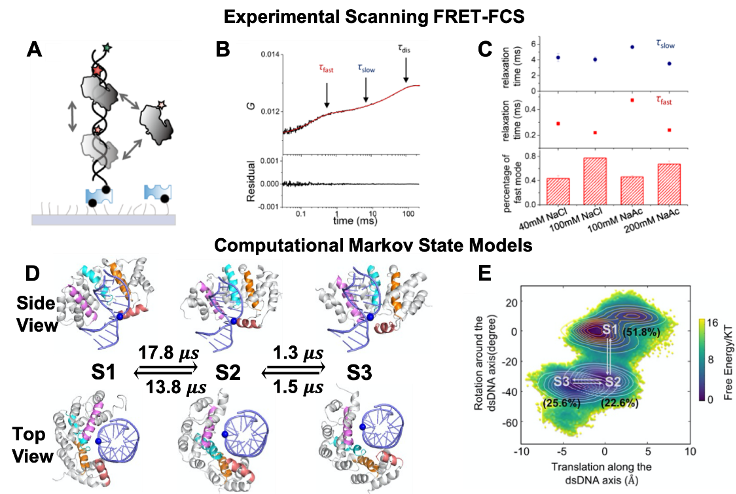
The researchers developed a scanning Fluorescence Resonance Energy Transfer (FRET) – Fluorescence Correlation Spectroscopy (FCS) platform to probe the protein dynamics at the microsecond temporal resolution and sub-nanometer spatial resolution. The significant improvement in the resolutions enables the researchers not only observe the fast mode (1D diffusion constant of ~ 8´106 bp2 s-1), but also directly capture the slow mode (1D diffusion constant of 6´104-5´105 bp2 s-1).
To further elucidate the underlying molecular mechanism of the slow mode, the researchers constructed the Markov State Model (MSM) based on extensive all-atom molecular dynamics (MD) simulations. Based on MSM, they visualized continuous cycles of AlkD diffusion along dsDNA over 1 ms, a timescale that is difficult to be reached by conventional MD simulations. They revealed that the diffusion of AlkD over one base pair contained a rate-limiting rotation and a sequential translation. Moreover, they pinpointed the essential role of Y27 in determining the AlkD diffusion dynamics both experimentally and computationally.
The study provided mechanistic insights on how conformational dynamics of AlkD-dsDNA complex coordinate different diffusion modes to recognize DNA lesions with high efficiency and accuracy. The mechanism adopted by AlkD to search for DNA lesions may be utilized by other glycosylases and DNA binding proteins. The integrated platform by combining scanning FRET-FCS with Markov State Model can be further widely applied to investigate other glycosylases and DNA-binding proteins.
Molecular Mechanisms of Target Search of AlkD Revealed by the Integrated
Platform Combining Scanning FRET-FCS and Markov State Model
This work is supported by the National Natural Science Foundation of China, Hong Kong Research Grant Council, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology and Beijing Frontier Research Center for Biological Structure.
Published Paper Title: Target search and recognition mechanisms of glycosylase AlkD revealed by scanning FRET-FCS and Markov state models
Link: https://doi.org/10.1073/pnas.2002971117
