On March 30, 2020, a research paper entitled “Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor” was published online in Nature as “Accelerated Article Preview” by the groups of Prof. Xinquan Wang at the School of Life Sciences and Prof. Linqi Zhang at the School of Medicine, Tsinghua University. This study reported the crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) in complex with the human cell receptor ACE2 at a resolution of 2.45 angstrom. By precisely elucidating the interactions between RBD and ACE2, this study revealed the structural basis for the recognition of SARS-CoV-2 by ACE2 and provided important structural knowledge for the development of antibodies and vaccines against the COVID-19 pandemic.
To help the research on the SARS-CoV-2 and anti-virus drug discovery, the paper has been deposited into the preprint server BioRxiv on February 20, 2020. The coordinates of the crystal structure were also available for download before the official release of the Protein Data Bank (PDB) on the website of the Beijing Advanced Innovation Center for Structural Biology (http://www.icsb.tsinghua.edu.cn).
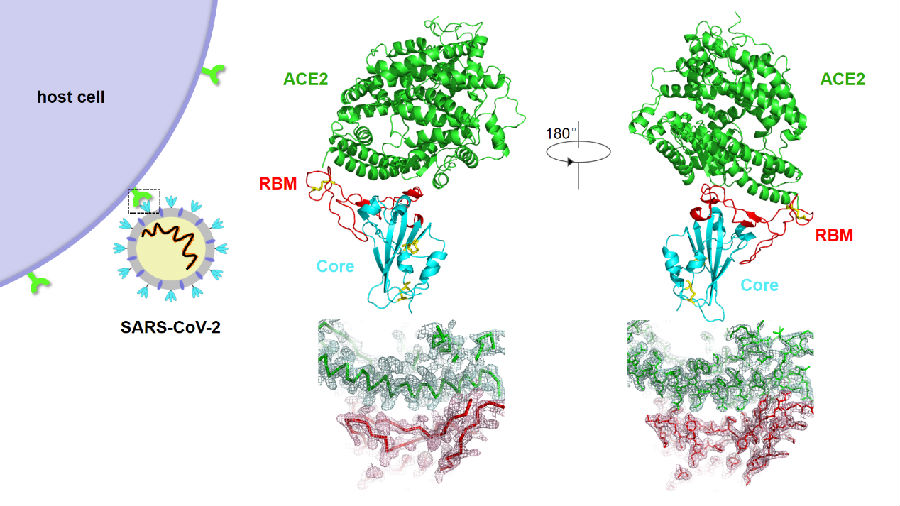
Crystal structure of the SARS-CoV-2 RBD (core+RBM) bound to the ACE2 receptor and the density map at the binding interface
To better understand the initial step of SARS-CoV-2 infection at atomic-level, the Wang and Zhang groups expressed and purified the SARS-CoV-2 RBD and human ACE2 in insect cells, crystallized the complex, and collected the diffraction dataset on the BL17U1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF). The overall ACE2-binding mode of the SARS-CoV-2 RBD is nearly identical to that of the SARS-CoV RBD, which also utilizes ACE2 as the cell receptor. Structural analysis identified residues in the SARS-CoV-2 RBD critical for ACE2 binding, and majority of which are either highly conserved or shared similar side chain properties with those in the SARS-CoV RBD. Such similarity in structure and sequence strongly argue for a convergent evolution between SARS-CoV-2 and SARS-CoV RBDs for improved binding to ACE2, although SARS-CoV-2 does not cluster within SARS and SARS-related coronaviruses. The epitopes of two SARS-CoV antibodies targeting the RBD are also analyzed with the SARS-CoV-2 RBD, providing insights into future identification of cross-reactive antibodies.
Ph.D. student Jun Lan, postdoc Jiwan Ge, PhD student Jinfang Yu from the School of Life Sciences and PhD student Sisi Shan from the School of Medicine are the co-first authors of the study. Dr. Shilong Fan from the Tsinghua University Technology Center for Protein Sciences, Drs Huan Zhou and Qisheng Wang from the SSRF, Drs. Xuanling Shi and Qi Zhang also contribute to the study. Professors Xinquan Wang and Linqi Zhang are the corresponding authors of the study. This study is supported by Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, MOE Key Laboratory of Protein Science, and the National Key Plan for Scientific Research and Development of China.
Article link: https://www.nature.com/articles/s41586-020-2180-5
